Difference between revisions of "FusA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' elongation factor G <br/><br/> | + | * '''Description:''' elongation factor G, facilitates movement of tRNA–mRNA by one codon <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || elongation factor G | |style="background:#ABCDEF;" align="center"| '''Product''' || elongation factor G | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || translation | + | |style="background:#ABCDEF;" align="center"|'''Function''' || [[translation]] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU01120 fusA] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU01120 fusA] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/FusA FusA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 76 kDa, 4.615 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 76 kDa, 4.615 | ||
| Line 35: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 93: | Line 91: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** FusA-RplL {{PubMed|23912278}} | ||
| + | ** FusA-RpsL {{PubMed|23912278}} | ||
* '''[[Localization]]:''' cytoplasm (according to Swiss-Prot) | * '''[[Localization]]:''' cytoplasm (according to Swiss-Prot) | ||
| Line 98: | Line 98: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?pdbId=1ELO 1ELO] (from ''Thermus thermophilus'') {{PubMed|8070397}} | + | * '''Structure:''' |
| + | ** [http://www.rcsb.org/pdb/explore/explore.do?pdbId=1ELO 1ELO] (from ''Thermus thermophilus'') {{PubMed|8070397}} | ||
| + | ** [http://www.pdb.org/pdb/explore/explore.do?structureId=4BTC 4BTC], [http://www.pdb.org/pdb/explore/explore.do?structureId=4BTD 4BTD], the ''Thermus thermophilus'' EF-G–[[ribosome]] complex in a pretranslocation state {{PubMed|23912278}} | ||
* '''UniProt:''' [http://www.uniprot.org/uniprot/P80868 P80868] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P80868 P80868] | ||
| Line 128: | Line 130: | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | ** for expression/ purification from ''B. subtilis'' with N-terminal Strep-tag, for [[SPINE]], in [[pGP380]]: pGP840, available in [[Stülke]] lab | + | ** for expression/ purification from ''B. subtilis'' with N-terminal Strep-tag, for [[SPINE]], in [[pGP380]]: pGP840, available in [[Jörg Stülke]]'s lab |
| − | ** for expression/ purification from ''E. coli'' with N-terminal His-tag, in [[pWH844]]: pGP848, available in [[Stülke]] lab | + | ** for expression/ purification from ''E. coli'' with N-terminal His-tag, in [[pWH844]]: pGP848, available in [[Jörg Stülke]]'s lab |
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| Line 145: | Line 147: | ||
=References= | =References= | ||
| − | <pubmed> 17726680, 17218307 20348921 8070397 </pubmed> | + | <pubmed> 17726680, 17218307 20348921 8070397 23912278</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:28, 20 August 2013
- Description: elongation factor G, facilitates movement of tRNA–mRNA by one codon
| Gene name | fusA |
| Synonyms | fus |
| Essential | yes PubMed |
| Product | elongation factor G |
| Function | translation |
| Gene expression levels in SubtiExpress: fusA | |
| Interactions involving this protein in SubtInteract: FusA | |
| MW, pI | 76 kDa, 4.615 |
| Gene length, protein length | 2076 bp, 692 aa |
| Immediate neighbours | rpsG, tufA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 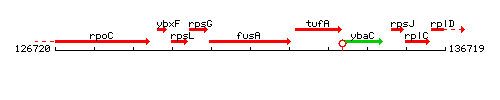
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
translation, essential genes, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01120
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: hydrolyses GTP
- Protein family: EF-G/EF-2 subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-213 AND Ser-302 AND Ser-569 AND Ser-680 AND (Thr-24 OR Thr-25) AND (Thr-43 OR Ser 48) PubMed, PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P80868
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP840, available in Jörg Stülke's lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP848, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References