Difference between revisions of "NagP"
| Line 28: | Line 28: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nagP_840656_842014_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:nagP_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nagP_840656_842014_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:nagP_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU07700]] |
|- | |- | ||
|} | |} | ||
Revision as of 12:46, 16 May 2013
- Description: N-acetylglucosamine-specific phosphotransferase system, EIICB of the PTS
| Gene name | nagP |
| Synonyms | yflF |
| Essential | no |
| Product | N-acetylglucosamine-specific PTS, EIICB |
| Function | N-acetylglucosamine uptake and phosphorylation |
| Gene expression levels in SubtiExpress: nagP | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 48 kDa, 7.127 |
| Gene length, protein length | 1356 bp, 452 aa |
| Immediate neighbours | yflG, ltaS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed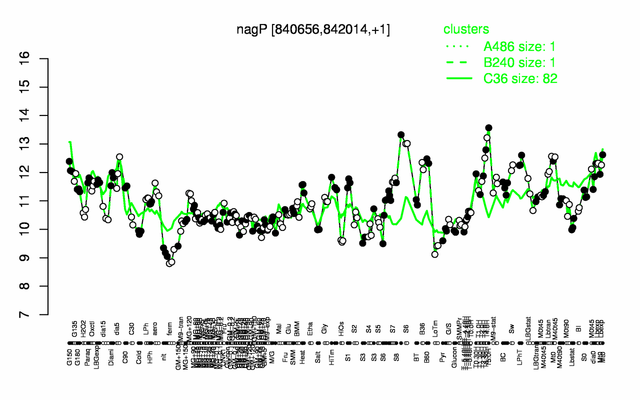
| |
Contents
Categories containing this gene/protein
phosphotransferase systems, utilization of specific carbon sources, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU07700
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot), membrane associated PubMed
Database entries
- Structure:
- UniProt: O34521
- KEGG entry: [2]
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)
H L Mobley, R J Doyle, U N Streips, S O Langemeier
Transport and incorporation of N-acetyl-D-glucosamine in Bacillus subtilis.
J Bacteriol: 1982, 150(1);8-15
[PubMed:6174502]
[WorldCat.org]
[DOI]
(P p)