Difference between revisions of "PbpC"
| Line 28: | Line 28: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pbpC_463934_465940_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:pbpC_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pbpC_463934_465940_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:pbpC_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU04140]] |
|- | |- | ||
|} | |} | ||
Revision as of 12:34, 16 May 2013
- Description: penicillin-binding protein 3
| Gene name | pbpC |
| Synonyms | ycsM, yzsA |
| Essential | no |
| Product | penicillin-binding protein 3 |
| Function | unknown |
| Gene expression levels in SubtiExpress: pbpC | |
| Interactions involving this protein in SubtInteract: PbpC | |
| MW, pI | 74 kDa, 6.202 |
| Gene length, protein length | 2004 bp, 668 aa |
| Immediate neighbours | yczJ, ycsN |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 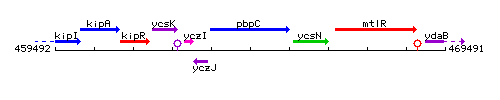
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed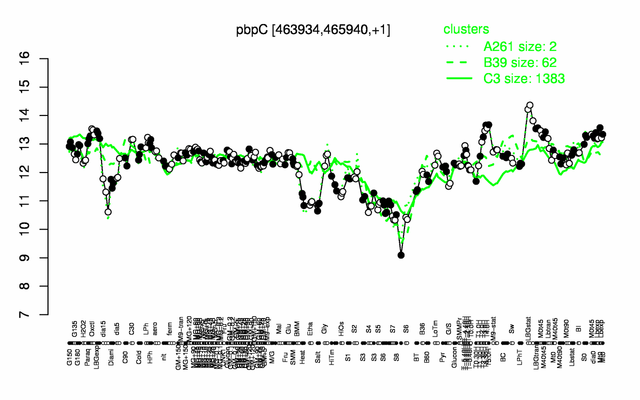
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04140
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: transpeptidase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- may be part of the cell wall biosynthetic complex PubMed
- folding requires PrsA PubMed
- Localization: cell membrane (according to Swiss-Prot), extracellular (signal peptide) PubMed
Database entries
- Structure:
- UniProt: P42971
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jeff Errington lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Hanne-Leena Hyyryläinen, Bogumila C Marciniak, Kathleen Dahncke, Milla Pietiäinen, Pascal Courtin, Marika Vitikainen, Raili Seppala, Andreas Otto, Dörte Becher, Marie-Pierre Chapot-Chartier, Oscar P Kuipers, Vesa P Kontinen
Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis.
Mol Microbiol: 2010, 77(1);108-27
[PubMed:20487272]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Dirk-Jan Scheffers, Laura J F Jones, Jeffery Errington
Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis.
Mol Microbiol: 2004, 51(3);749-64
[PubMed:14731276]
[WorldCat.org]
[DOI]
(P p)
T Murray, D L Popham, P Setlow
Identification and characterization of pbpC, the gene encoding Bacillus subtilis penicillin-binding protein 3.
J Bacteriol: 1996, 178(20);6001-5
[PubMed:8830698]
[WorldCat.org]
[DOI]
(P p)