Difference between revisions of "PckA"
| Line 26: | Line 26: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[metK]]'', ''[[ytmB]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[metK]]'', ''[[ytmB]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU30560 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU30560 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU30560 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU30560 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU30560 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU30560 DNA_with_flanks] |
|- | |- | ||
|- | |- | ||
Revision as of 11:04, 14 May 2013
- Description: phosphoenolpyruvate carboxykinase
| Gene name | pckA |
| Synonyms | ppc |
| Essential | no |
| Product | phosphoenolpyruvate carboxykinase |
| Function | synthesis of phosphoenolpyruvate |
| Gene expression levels in SubtiExpress: pckA | |
| Interactions involving this protein in SubtInteract: PckA | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 58,1 kDa, 5.12 |
| Gene length, protein length | 1581 bp, 527 amino acids |
| Immediate neighbours | metK, ytmB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 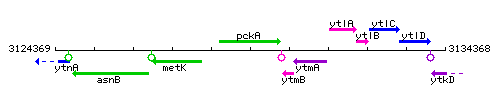
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed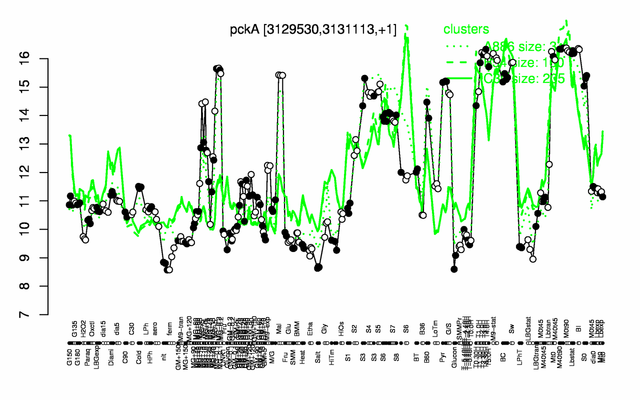
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30560
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO2 (according to Swiss-Prot) ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO(2)
- Protein family: phosphoenolpyruvate carboxykinase [ATP] family (according to Swiss-Prot) phosphoenolpyruvate carboxykinase [ATP] family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Nucleotide binding Domain (233–240)
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot), cytoplasm
Database entries
- Structure: 2PXZ (E.coli)
- UniProt: P54418
- KEGG entry: [3]
- E.C. number: 4.1.1.49
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- 1A1005 ( pckA::spec), PubMed, available at BGSC
- 1A996 ( pckA::spec), PubMed, available at BGSC
- GP1147 (pckA::neo), available in Jörg Stülke's lab
- Expression vector:
- pGP1753 (N-terminal Strep-tag, for SPINE, purification from B. subtilis, in pGP380) (available in Jörg Stülke's lab)
- pGP1762 (for expression, purification in E. coli with N-terminal His-tag, in pWH844, available in Jörg Stülke's lab)
- pGP1763 (for expression, purification in E. coli with N-terminal Strep-tag, in pGP172, available in Jörg Stülke's lab)
- lacZ fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
- FLAG-tag construct:
- GP1129 (spc, based on pGP1331), available in Jörg Stülke's lab
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References