Difference between revisions of "AlsR"
| Line 81: | Line 81: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| + | ** K(D) value for the binding site in the ''[[alsS]]'' promoter region: 33.5 nM {{PubMed|22178965}} | ||
* '''Domains:''' | * '''Domains:''' | ||
| Line 112: | Line 113: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=alsR_3711498_3712406_1 alsR] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=alsR_3711498_3712406_1 alsR] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 19:48, 31 January 2013
| Gene name | alsR |
| Synonyms | |
| Essential | no |
| Product | transcriptional activator(LysR family) |
| Function | regulation of acetoin synthesis (alsS-alsD) |
| Gene expression levels in SubtiExpress: alsR | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 34 kDa, 7.288 |
| Gene length, protein length | 906 bp, 302 aa |
| Immediate neighbours | alsS, ywrK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 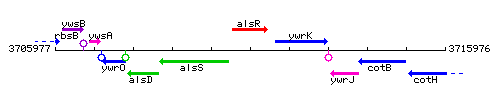
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed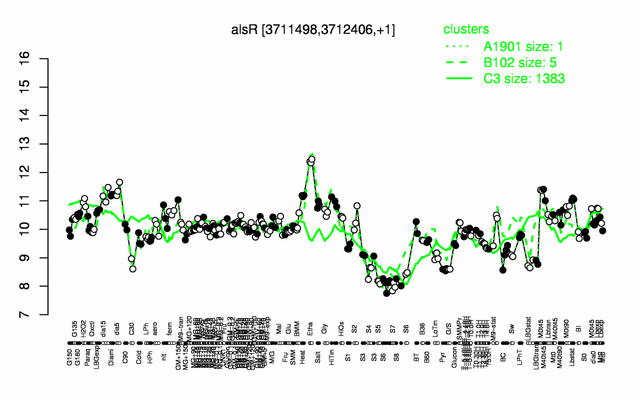
| |
Contents
Categories containing this gene/protein
carbon core metabolism, transcription factors and their control
This gene is a member of the following regulons
The AlsR regulon:
The gene
Basic information
- Locus tag: BSU36020
Phenotypes of a mutant
no acetoin production
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
- Inactivation of alsR, sigD and sigW increases competitive fitness of Bacillus subtilis under non-sporulating conditions PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: binds the promoter region of the alsS-alsD operon in the presence of acetate, activates transcription
- Protein family: LysR family of transcription factors
- Paralogous protein(s):
Extended information on the protein
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q04778
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Elisabeth Härtig, Braunschweig, Germany Homepage
Your additional remarks
References
Reviews
Elisabeth Härtig, Dieter Jahn
Regulation of the anaerobic metabolism in Bacillus subtilis.
Adv Microb Physiol: 2012, 61;195-216
[PubMed:23046954]
[WorldCat.org]
[DOI]
(I p)
Original publications
Additional publications: PubMed
Wayne L Nicholson
Increased competitive fitness of Bacillus subtilis under nonsporulating conditions via inactivation of pleiotropic regulators AlsR, SigD, and SigW.
Appl Environ Microbiol: 2012, 78(9);3500-3
[PubMed:22344650]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
M C Renna, N Najimudin, L R Winik, S A Zahler
Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin.
J Bacteriol: 1993, 175(12);3863-75
[PubMed:7685336]
[WorldCat.org]
[DOI]
(P p)