Difference between revisions of "Spx"
(→References) |
|||
| Line 38: | Line 38: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
Revision as of 14:38, 18 September 2012
- Description: transcriptional regulator Spx, involved in regulation of many genes.
| Gene name | spx |
| Synonyms | yjbD |
| Essential | no |
| Product | transcriptional regulator Spx |
| Function | negative and positive regulator of many genes |
| Gene expression levels in SubtiExpress: spx | |
| Interactions involving this protein in SubtInteract: Spx | |
| Metabolic function and regulation of this protein in SubtiPathways: Riboflavin / FAD | |
| MW, pI | 15,5 kDa, 7.80 |
| Gene length, protein length | 393 bp, 131 amino acids |
| Immediate neighbours | yjbC, yjbE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 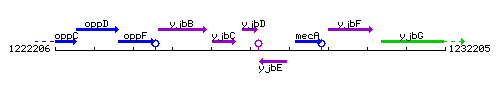
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription factors and their control, general stress proteins (controlled by SigB), cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
PerR regulon, SigB regulon, SigM regulon, SigW regulon, SigX regulon
The Spx regulon
The gene
Basic information
- Locus tag: BSU11500
Phenotypes of a mutant
- Loss of up-regulation of the methionine sulfoxide reductase (msrA-msrB) operon in response to thiol specific oxidative stress, also loss of trxA and trxB upregulation in response to thiol specific oxidative stress.
Database entries
- DBTBS entry: [1]
- SubtiList entry: link
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- transcriptional regulator of many genes in response to thiol specific oxidative stress (transcription activator of trxA and trxB)
- in addition, Spx inhibits transcription by binding to the C-terminal domain of the alpha subunit of RNAP (RpoA), disrupting complex formation between RNAP and certain transcriptional activator proteins like ResD and ComA
- in response to thiol specific oxidative stress, Spx can also activate transcription, making it a general regulator that exerts both positive and negative control over transcription initiation
- involved in competence regulation PubMed
- Protein family: Spx subfamily (according to Swiss-Prot) Arsenate Reductase (ArsC) family, Spx subfamily
- Paralogous protein(s): MgsR
Extended information on the protein
- Kinetic information:
- Domains: CXXC (10-13): Acts as a disulfide switch for the redox-sensitive transcriptional regulation of genes that function in thiol homeostasis.
- Modification: Cysteine oxidation of the CXXC motif
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: O31602
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Additional information:
- post-translational control by ClpX-ClpP: Spx naturally contains a C-terminal sequence that resembles the SsrA tag and targets the protein for degradation. PubMed
- proteolysis is enhanced by YjbH PubMed and counter-acted by YirB PubMed
- the mRNA is substantially stabilized upon depletion of RNase Y (the half-life of the monocistronic spx mRNA increases from 1 to 6 min) PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Peter Zuber, Oregon Health and Science University, USA Homepage
Richard Brennan, Houston, Texas, USA Homepage
Your additional remarks
References
Reviews
Additional reviews: PubMed
The Spx regulon
Additional publications: PubMed
Structural analysis of Spx
Original Publications
Additional publications: PubMed
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
Michiko M Nakano, Ann Lin, Cole S Zuber, Kate J Newberry, Richard G Brennan, Peter Zuber
Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit.
PLoS One: 2010, 5(1);e8664
[PubMed:20084284]
[WorldCat.org]
[DOI]
(I e)
Andriansjah Rukmana, Takuya Morimoto, Hiroki Takahashi, Giyanto, Naotake Ogasawara
Assessment of transcriptional responses of Bacillus subtilis cells to the antibiotic enduracidin, which interferes with cell wall synthesis, using a high-density tiling chip.
Genes Genet Syst: 2009, 84(4);253-67
[PubMed:20057163]
[WorldCat.org]
[DOI]
(P p)
Saurabh K Garg, Sushma Kommineni, Luke Henslee, Ying Zhang, Peter Zuber
The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx.
J Bacteriol: 2009, 191(4);1268-77
[PubMed:19074380]
[WorldCat.org]
[DOI]
(I p)
Dindo Y Reyes, Peter Zuber
Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation.
Mol Microbiol: 2008, 69(3);765-79
[PubMed:18687074]
[WorldCat.org]
[DOI]
(I p)
CongHui You, Agnieszka Sekowska, Olivera Francetic, Isabelle Martin-Verstraete, YiPing Wang, Antoine Danchin
Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis.
BMC Microbiol: 2008, 8;128
[PubMed:18662407]
[WorldCat.org]
[DOI]
(I e)
Falko Hochgräfe, Carmen Wolf, Stephan Fuchs, Manuel Liebeke, Michael Lalk, Susanne Engelmann, Michael Hecker
Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus.
J Bacteriol: 2008, 190(14);4997-5008
[PubMed:18487332]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Jonas T Larsson, Annika Rogstam, Claes von Wachenfeldt
YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis.
Mol Microbiol: 2007, 66(3);669-84
[PubMed:17908206]
[WorldCat.org]
[DOI]
(P p)
Ying Zhang, Peter Zuber
Requirement of the zinc-binding domain of ClpX for Spx proteolysis in Bacillus subtilis and effects of disulfide stress on ClpXP activity.
J Bacteriol: 2007, 189(21);7669-80
[PubMed:17827297]
[WorldCat.org]
[DOI]
(P p)
Adrian J Jervis, Penny D Thackray, Chris W Houston, Malcolm J Horsburgh, Anne Moir
SigM-responsive genes of Bacillus subtilis and their promoters.
J Bacteriol: 2007, 189(12);4534-8
[PubMed:17434969]
[WorldCat.org]
[DOI]
(P p)
Montira Leelakriangsak, Kazuo Kobayashi, Peter Zuber
Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis.
J Bacteriol: 2007, 189(5);1736-44
[PubMed:17158660]
[WorldCat.org]
[DOI]
(P p)
Soon-Yong Choi, Dindo Reyes, Montira Leelakriangsak, Peter Zuber
The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis.
J Bacteriol: 2006, 188(16);5741-51
[PubMed:16885442]
[WorldCat.org]
[DOI]
(P p)
Ying Zhang, Shunji Nakano, Soon-Yong Choi, Peter Zuber
Mutational analysis of the Bacillus subtilis RNA polymerase alpha C-terminal domain supports the interference model of Spx-dependent repression.
J Bacteriol: 2006, 188(12);4300-11
[PubMed:16740936]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Shunji Nakano, Kyle N Erwin, Martina Ralle, Peter Zuber
Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx.
Mol Microbiol: 2005, 55(2);498-510
[PubMed:15659166]
[WorldCat.org]
[DOI]
(P p)
Penny D Thackray, Anne Moir
SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress.
J Bacteriol: 2003, 185(12);3491-8
[PubMed:12775685]
[WorldCat.org]
[DOI]
(P p)
Shunji Nakano, Michiko M Nakano, Ying Zhang, Montira Leelakriangsak, Peter Zuber
A regulatory protein that interferes with activator-stimulated transcription in bacteria.
Proc Natl Acad Sci U S A: 2003, 100(7);4233-8
[PubMed:12642660]
[WorldCat.org]
[DOI]
(P p)
Shunji Nakano, Guolu Zheng, Michiko M Nakano, Peter Zuber
Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis.
J Bacteriol: 2002, 184(13);3664-70
[PubMed:12057962]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, F Hajarizadeh, Y Zhu, P Zuber
Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis.
Mol Microbiol: 2001, 42(2);383-94
[PubMed:11703662]
[WorldCat.org]
[DOI]
(P p)
H Antelmann, C Scharf, M Hecker
Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis.
J Bacteriol: 2000, 182(16);4478-90
[PubMed:10913081]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, J Bernhardt, U Gerth, D Höper, T Koburger, U Völker, M Hecker
Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization.
J Bacteriol: 1999, 181(18);5718-24
[PubMed:10482513]
[WorldCat.org]
[DOI]
(P p)