Difference between revisions of "RsiX"
| Line 34: | Line 34: | ||
__TOC__ | __TOC__ | ||
| − | |||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| Line 59: | Line 55: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * inactivation of ''[[rsiX]]'' strongly restores beta-lactam resistance in a ''[[sigM]]'' mutant {{PubMed|22211522}} | ||
=== Database entries === | === Database entries === | ||
| Line 67: | Line 64: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 143: | Line 137: | ||
=References= | =References= | ||
| − | + | '''Additional publications:''' {{PubMed|22211522}} | |
<pubmed>14993308,7934830, 16306698 9636707 9393439 </pubmed> | <pubmed>14993308,7934830, 16306698 9636707 9393439 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:07, 21 July 2012
- Description: anti-SigX
| Gene name | rsiX |
| Synonyms | ypuN |
| Essential | no |
| Product | anti-SigX |
| Function | control of SigX activity |
| Interactions involving this protein in SubtInteract: RsiX | |
| MW, pI | 41 kDa, 5.452 |
| Gene length, protein length | 1104 bp, 368 aa |
| Immediate neighbours | aroC, sigX |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 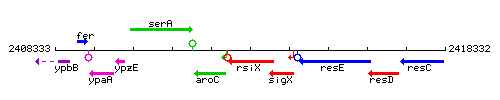
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed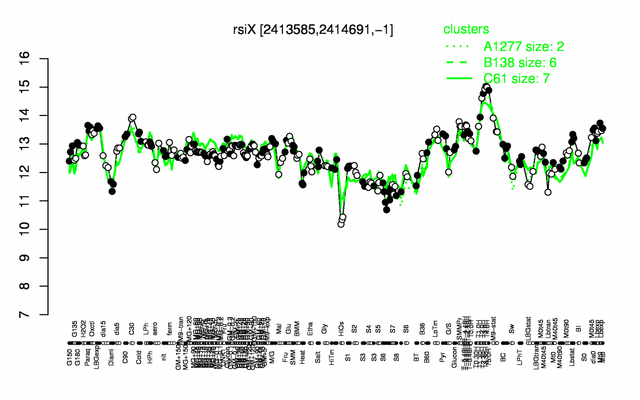
| |
Contents
Categories containing this gene/protein
sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23090
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane
Database entries
- Structure:
- UniProt: P35166
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
Mika Yoshimura, Kei Asai, Yoshito Sadaie, Hirofumi Yoshikawa
Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors.
Microbiology (Reading): 2004, 150(Pt 3);591-599
[PubMed:14993308]
[WorldCat.org]
[DOI]
(P p)
X Huang, J D Helmann
Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search.
J Mol Biol: 1998, 279(1);165-73
[PubMed:9636707]
[WorldCat.org]
[DOI]
(P p)
S Brutsche, V Braun
SigX of Bacillus subtilis replaces the ECF sigma factor fecI of Escherichia coli and is inhibited by RsiX.
Mol Gen Genet: 1997, 256(4);416-25
[PubMed:9393439]
[WorldCat.org]
[DOI]
(P p)
V Azevedo, A Sorokin, S D Ehrlich, P Serror
The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci.
Mol Microbiol: 1993, 10(2);397-405
[PubMed:7934830]
[WorldCat.org]
[DOI]
(P p)