Difference between revisions of "NrnA"
| Line 104: | Line 104: | ||
* '''Operon:''' ''[[nrnA]]'' {{PubMed|9387221}} | * '''Operon:''' ''[[nrnA]]'' {{PubMed|9387221}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nrnA_2995908_2996849_-1 nrnA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 110: | Line 112: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 15:10, 16 April 2012
Description: oligoribonuclease (nanoRNase), 3',5'-bisphosphate nucleotidase
| Gene name | nrnA |
| Synonyms | ytqI |
| Essential | no |
| Product | oligoribonuclease (nanoRNase), 3',5'-bisphosphate nucleotidase |
| Function | degradation of RNA oligonucleotides, |
| MW, pI | 34 kDa, 4.668 |
| Gene length, protein length | 939 bp, 313 aa |
| Immediate neighbours | ytzJ, ytpI |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 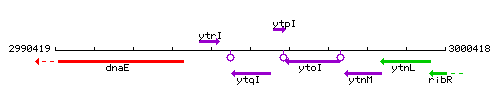
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29250
Phenotypes of a mutant
The mutant requires cysteine PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- Cytoplasm (Homogeneous) PubMed
Database entries
- Structure:
- UniProt: O34600
- KEGG entry: [3]
- E.C. number: 3.1.3.7
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Taisuke Wakamatsu, Kwang Kim, Yuri Uemura, Noriko Nakagawa, Seiki Kuramitsu, Ryoji Masui
Role of RecJ-like protein with 5'-3' exonuclease activity in oligo(deoxy)nucleotide degradation.
J Biol Chem: 2011, 286(4);2807-16
[PubMed:21087930]
[WorldCat.org]
[DOI]
(I p)
Ming Fang, Wencke-Maria Zeisberg, Ciaran Condon, Vasily Ogryzko, Antoine Danchin, Undine Mechold
Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis.
Nucleic Acids Res: 2009, 37(15);5114-25
[PubMed:19553197]
[WorldCat.org]
[DOI]
(I p)
Jiaqin Zhang, Indranil Biswas
3'-Phosphoadenosine-5'-phosphate phosphatase activity is required for superoxide stress tolerance in Streptococcus mutans.
J Bacteriol: 2009, 191(13);4330-40
[PubMed:19429620]
[WorldCat.org]
[DOI]
(I p)
Undine Mechold, Gang Fang, Saravuth Ngo, Vasily Ogryzko, Antoine Danchin
YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity.
Nucleic Acids Res: 2007, 35(13);4552-61
[PubMed:17586819]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)