Difference between revisions of "CypC"
(→References) |
(→References) |
||
| Line 128: | Line 128: | ||
=References= | =References= | ||
'''Additional publications:''' {{PubMed|17385817,11914497}} | '''Additional publications:''' {{PubMed|17385817,11914497}} | ||
| − | <pubmed>,10529095,12519760,15805528, 20697922 | + | <pubmed>12297285 11827534 11566026 17385817,11914497</pubmed> |
| + | --- | ||
| + | <pubmed>,10529095,12519760,15805528, 20697922 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:21, 16 April 2011
- Description: long chain-fatty acid beta-hydroxylating cytochrome P450, H(2)O(2)-dependent, hydroxylates myristic acid to beta-hydroxymyristic acid
| Gene name | cypC |
| Synonyms | ybdT |
| Essential | no |
| Product | fatty acid beta-hydroxylating cytochrome P450 |
| Function | biosynthesis of beta-hydroxy fatty acid |
| MW, pI | 47 kDa, 6.468 |
| Gene length, protein length | 1251 bp, 417 aa |
| Immediate neighbours | ybxI, ybyB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 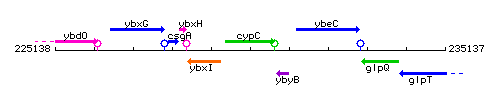
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
electron transport/ other, lipid metabolism/ other
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- hydroxylation of a long-chain fatty acid (e.g. myristic acid) at the alpha- and beta-positions using hydrogen peroxide as an oxidant PubMed
- Protein family: cytochrome P450 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: O31440
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: cypC (according to DBTBS)
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Osami Shoji, Takashi Fujishiro, Hiroshi Nakajima, Misa Kim, Shingo Nagano, Yoshitsugu Shiro, Yoshihito Watanabe
Hydrogen peroxide dependent monooxygenations by tricking the substrate recognition of cytochrome P450BSbeta.
Angew Chem Int Ed Engl: 2007, 46(20);3656-9
[PubMed:17385817]
[WorldCat.org]
[DOI]
(P p)
Isamu Matsunaga, Tatsuo Sumimoto, Minoru Ayata, Hisashi Ogura
Functional modulation of a peroxygenase cytochrome P450: novel insight into the mechanisms of peroxygenase and peroxidase enzymes.
FEBS Lett: 2002, 528(1-3);90-4
[PubMed:12297285]
[WorldCat.org]
[DOI]
(P p)
Dong Sun Lee, Akari Yamada, Isamu Matsunaga, Kosuke Ichihara, Shin-ichi Adachi, Sam-Yong Park, Yoshitsugu Shiro
Crystallization and preliminary X-ray diffraction analysis of fatty-acid hydroxylase cytochrome P450BSbeta from Bacillus subtilis.
Acta Crystallogr D Biol Crystallogr: 2002, 58(Pt 4);687-9
[PubMed:11914497]
[WorldCat.org]
[DOI]
(P p)
Isamu Matsunaga, Akari Yamada, Dong-Sun Lee, Eiji Obayashi, Nagatoshi Fujiwara, Kazuo Kobayashi, Hisashi Ogura, Yoshitsugu Shiro
Enzymatic reaction of hydrogen peroxide-dependent peroxygenase cytochrome P450s: kinetic deuterium isotope effects and analyses by resonance Raman spectroscopy.
Biochemistry: 2002, 41(6);1886-92
[PubMed:11827534]
[WorldCat.org]
[DOI]
(P p)
I Matsunaga, A Ueda, T Sumimoto, K Ichihara, M Ayata, H Ogura
Site-directed mutagenesis of the putative distal helix of peroxygenase cytochrome P450.
Arch Biochem Biophys: 2001, 394(1);45-53
[PubMed:11566026]
[WorldCat.org]
[DOI]
(P p)
---