Difference between revisions of "Mbl"
| Line 57: | Line 57: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 114: | Line 111: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** An [[ncRNA|antisense RNA]] is predicted for ''[[mbl]]'' {{PubMed|20525796}} | ||
=Biological materials = | =Biological materials = | ||
| Line 139: | Line 137: | ||
<pubmed> 17064365, 20566861 </pubmed> | <pubmed> 17064365, 20566861 </pubmed> | ||
==Other original publications== | ==Other original publications== | ||
| − | <pubmed>19643765,16832063,11290328,12809607,16101995,12530960,7836311,,16950129 9023218, 19659933 19114499, 21091501</pubmed> | + | <pubmed>19643765,16832063,11290328,12809607,16101995,12530960,7836311,,16950129 9023218, 19659933 19114499, 21091501 20525796</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:39, 4 May 2011
- Description: cell-shape determining protein, forms filaments
| Gene name | mbl |
| Synonyms | |
| Essential | no |
| Product | MreB-like protein |
| Function | cell-shape determination |
| MW, pI | 35 kDa, 5.669 |
| Gene length, protein length | 999 bp, 333 aa |
| Immediate neighbours | flhO, spoIIID |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 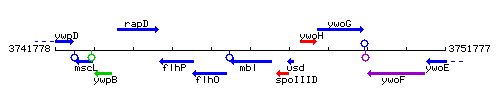
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
cell shape, sporulation proteins, membrane proteins
This gene is a member of the following regulons
SigE regulon, stringent response
The gene
Basic information
- Locus tag: BSU36410
Phenotypes of a mutant
non-viable in the presence of low Mg(2+), readily accumulate rsgI suppressor mutants PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- forms helical filaments in a heterologous system PubMed
- Protein family: ftsA/mreB family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P39751
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- An antisense RNA is predicted for mbl PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Görke lab
- Antibody: available in the Jeff Errington and Peter Graumann labs
Labs working on this gene/protein
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Localization
Henrik Strahl, Leendert W Hamoen
Membrane potential is important for bacterial cell division.
Proc Natl Acad Sci U S A: 2010, 107(27);12281-6
[PubMed:20566861]
[WorldCat.org]
[DOI]
(I p)
Hervé Joël Defeu Soufo, Peter L Graumann
Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB.
Mol Microbiol: 2006, 62(5);1340-56
[PubMed:17064365]
[WorldCat.org]
[DOI]
(P p)
Other original publications