Difference between revisions of "Easter egg"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' RNA component of [[RNase]] P <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''rnpB'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''rnaP '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || RNA component of ribonuclease P <br/>([[RNase]] P) (catalytic subunit, ribozyme) |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || cleavage of precursor sequences <br/>from the 5' ends of pre-tRNAs |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length''' || 401 bp |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ypsC]]'', ''[[ypsB]]'' |
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/rnpB_nucleotide.txt Gene sequence (+200bp) ]''' |
| + | |||
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:rnpB_context.gif]] |
| + | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
|} | |} | ||
| Line 26: | Line 28: | ||
__TOC__ | __TOC__ | ||
| − | <br/><br/> | + | <br/><br/><br/><br/><br/><br/><br/> |
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[ | + | {{SubtiWiki category|[[Rnases]]}}, |
| + | {{SubtiWiki category|[[translation]]}}, | ||
{{SubtiWiki category|[[ncRNA]]}} | {{SubtiWiki category|[[ncRNA]]}} | ||
| Line 44: | Line 46: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | === Database entries === | |
| + | |||
| + | * '''DBTBS entry:''' no entry | ||
| + | |||
| + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG00045] | ||
| + | |||
| + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2A64 2A64] (from Geobacillus stearothermophilus), [http://www.rcsb.org/pdb/explore.do?structureId=1NBS 1NBS] (specificity domain) | ||
| − | |||
| − | + | =The RNA= | |
| − | * ''' | + | * '''Biological activity:''' 5' end maturation of precursor tRNAs |
| − | |||
| − | |||
| − | + | * '''RNA family:''' | |
| + | * '''cofactor:''' divalent cations (Ca(II) or Mg (II)) for the stabilization of the complex with pre-tRNA {{PubMed|20434461}} | ||
| + | * '''interaction:''' [[RnpA]]-RnpB | ||
| + | === Additional information=== | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' |
| − | * '''Sigma factor:''' | + | * '''Sigma factor:''' |
| − | * '''Regulation:''' | + | * '''Regulation:''' |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 80: | Line 88: | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| + | * '''GFP fusion:''' | ||
| − | + | * '''two-hybrid system:''' | |
| − | + | * '''Antibody:''' | |
| − | [[Roland Hartmann]] | + | =Labs working on this gene/RNA= |
| + | * [[Carol Fierke]], University of Michigan, Ann Arbor, USA [https://www.chem.lsa.umich.edu/chem/faculty/facultyDetail.php?Uniqname=fierke homepage] | ||
| + | * [[Roland Hartmann]], Marburg University, Germany [http://www.pharmazie.uni-marburg.de/pharmchem/akhartmann/ag_hartmann.htm homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
=References= | =References= | ||
| + | ==Reviews== | ||
| + | <pubmed> 19215776 17868095 14691942 16650980 16980936 16679018 14550630 </pubmed> | ||
| − | <pubmed> | + | ==Original Publications== |
| + | <pubmed>16470227,17355991,11812129,10390342,11454206, 19549719,11454206, 19717440 ,20133747 19932118 20188108 20434461 17299131 14510496 12951523 12836338 11577704 20557101 </pubmed> | ||
[[Category:small RNA]] | [[Category:small RNA]] | ||
Revision as of 13:50, 8 December 2010
- Description: RNA component of RNase P
| Gene name | rnpB |
| Synonyms | rnaP |
| Essential | no |
| Product | RNA component of ribonuclease P (RNase P) (catalytic subunit, ribozyme) |
| Function | cleavage of precursor sequences from the 5' ends of pre-tRNAs |
| Gene length | 401 bp |
| Immediate neighbours | ypsC, ypsB |
| Gene sequence (+200bp) | |
Genetic context 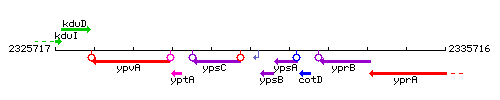
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
The RNA
- Biological activity: 5' end maturation of precursor tRNAs
- RNA family:
- cofactor: divalent cations (Ca(II) or Mg (II)) for the stabilization of the complex with pre-tRNA PubMed
- interaction: RnpA-RnpB
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/RNA
- Carol Fierke, University of Michigan, Ann Arbor, USA homepage
- Roland Hartmann, Marburg University, Germany homepage
Your additional remarks
References
Reviews
Original Publications
Ruiting Liang, Elzbieta Kierzek, Ryszard Kierzek, Douglas H Turner
Comparisons between chemical mapping and binding to isoenergetic oligonucleotide microarrays reveal unexpected patterns of binding to the Bacillus subtilis RNase P RNA specificity domain.
Biochemistry: 2010, 49(37);8155-68
[PubMed:20557101]
[WorldCat.org]
[DOI]
(I p)
John Hsieh, Kristin S Koutmou, David Rueda, Markos Koutmos, Nils G Walter, Carol A Fierke
A divalent cation stabilizes the active conformation of the B. subtilis RNase P x pre-tRNA complex: a role for an inner-sphere metal ion in RNase P.
J Mol Biol: 2010, 400(1);38-51
[PubMed:20434461]
[WorldCat.org]
[DOI]
(I p)
Nathan J Baird, Haipeng Gong, Syed S Zaheer, Karl F Freed, Tao Pan, Tobin R Sosnick
Extended structures in RNA folding intermediates are due to nonnative interactions rather than electrostatic repulsion.
J Mol Biol: 2010, 397(5);1298-306
[PubMed:20188108]
[WorldCat.org]
[DOI]
(I p)
Kristin S Koutmou, Anette Casiano-Negroni, Melissa M Getz, Samuel Pazicni, Andrew J Andrews, James E Penner-Hahn, Hashim M Al-Hashimi, Carol A Fierke
NMR and XAS reveal an inner-sphere metal binding site in the P4 helix of the metallo-ribozyme ribonuclease P.
Proc Natl Acad Sci U S A: 2010, 107(6);2479-84
[PubMed:20133747]
[WorldCat.org]
[DOI]
(I p)
Kristin S Koutmou, Nathan H Zahler, Jeffrey C Kurz, Frank E Campbell, Michael E Harris, Carol A Fierke
Protein-precursor tRNA contact leads to sequence-specific recognition of 5' leaders by bacterial ribonuclease P.
J Mol Biol: 2010, 396(1);195-208
[PubMed:19932118]
[WorldCat.org]
[DOI]
(I p)
Stefanie A Mortimer, Kevin M Weeks
C2'-endo nucleotides as molecular timers suggested by the folding of an RNA domain.
Proc Natl Acad Sci U S A: 2009, 106(37);15622-7
[PubMed:19717440]
[WorldCat.org]
[DOI]
(I p)
John Hsieh, Carol A Fierke
Conformational change in the Bacillus subtilis RNase P holoenzyme--pre-tRNA complex enhances substrate affinity and limits cleavage rate.
RNA: 2009, 15(8);1565-77
[PubMed:19549719]
[WorldCat.org]
[DOI]
(I p)
Barbara Wegscheid, Roland K Hartmann
In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3'-CCA.
Nucleic Acids Res: 2007, 35(6);2060-73
[PubMed:17355991]
[WorldCat.org]
[DOI]
(I p)
Somashekarappa Niranjanakumari, Jeremy J Day-Storms, Mahiuddin Ahmed, John Hsieh, Nathan H Zahler, Ronald A Venters, Carol A Fierke
Probing the architecture of the B. subtilis RNase P holoenzyme active site by cross-linking and affinity cleavage.
RNA: 2007, 13(4);521-35
[PubMed:17299131]
[WorldCat.org]
[DOI]
(P p)
Barbara Wegscheid, Ciarán Condon, Roland K Hartmann
Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences.
EMBO Rep: 2006, 7(4);411-7
[PubMed:16470227]
[WorldCat.org]
[DOI]
(P p)
Tomoaki Ando, Terumichi Tanaka, Yo Kikuchi
Bacterial ribonuclease P reaction is affected by substrate shape and magnesium ion concentration.
Nucleic Acids Res Suppl: 2003, (3);293-4
[PubMed:14510496]
[DOI]
(P p)
Tomoaki Ando, Terumichi Tanaka, Yo Kikuchi
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ribozymes.
Biosci Biotechnol Biochem: 2003, 67(8);1825-7
[PubMed:12951523]
[WorldCat.org]
[DOI]
(P p)
Y Hori, T Tanaka, Y Kikuchi
In vitro hyperprocessing of tRNAs by Bacillus subtilis ribonuclease P RNA.
Nucleic Acids Res Suppl: 2001, (1);209-10
[PubMed:12836338]
[DOI]
(P p)
Christoph Rox, Ralph Feltens, Thomas Pfeiffer, Roland K Hartmann
Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme.
J Mol Biol: 2002, 315(4);551-60
[PubMed:11812129]
[WorldCat.org]
[DOI]
(P p)
Y Hori, E Sakai, T Tanaka, Y Kikuchi
Hyperprocessing reaction of tRNA by Bacillus subtilis ribonuclease P ribozyme.
FEBS Lett: 2001, 505(2);337-9
[PubMed:11577704]
[WorldCat.org]
[DOI]
(P p)
A Hansen, T Pfeiffer, T Zuleeg, S Limmer, J Ciesiolka, R Feltens, R K Hartmann
Exploring the minimal substrate requirements for trans-cleavage by RNase P holoenzymes from Escherichia coli and Bacillus subtilis.
Mol Microbiol: 2001, 41(1);131-43
[PubMed:11454206]
[WorldCat.org]
[DOI]
(P p)
J M Warnecke, R Held, S Busch, R K Hartmann
Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis.
J Mol Biol: 1999, 290(2);433-45
[PubMed:10390342]
[WorldCat.org]
[DOI]
(P p)