Difference between revisions of "CggR"
| Line 33: | Line 33: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[carbon core metabolism]]}}, | ||
| + | {{SubtiWiki category|[[regulators of core metabolism]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[CggR regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 58: | ||
| − | + | ||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 23:28, 8 December 2010
- Description: repressor of the glycolytic gapA operon, DeoR family
| Gene name | cggR |
| Synonyms | yvbQ |
| Essential | no |
| Product | central glycolytic genes regulator |
| Function | transcriptional regulator |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 37,2 kDa,5.68 |
| Gene length, protein length | 1020 bp, 340 amino acids |
| Immediate neighbours | gapA, araE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 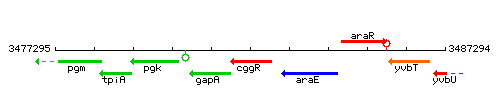
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
carbon core metabolism, regulators of core metabolism
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33950
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription repression of the glycolytic gapA operon
- Protein family: sorC transcriptional regulatory family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- DNA binding domain (H-T-H motif) (37–56)
- Modification:
- Cofactor(s):
- Effectors of protein activity: fructose 1.6-bisphosphate PubMed and dihydroxyacetone phosphate, glucose-6-phosphate and fructose-6-phosphate PubMed act as inducer and result in release of CggR from the DNA
- Interactions:
- active as dimer (according to PubMed)
- Localization: cytoplasma PubMed
Database entries
- Structure: 2OKG ( effector binding domain), 3BXH (in complex with fructose-6-phosphate), complex with Fructose-6-Phosphate NCBI, effector binding domain NCBI
- UniProt: O32253
- KEGG entry: [3]
Additional information
Expression and regulation
- Database entries: DBTBS
- Additional information:
- The primary mRNAs of the operon are highly unstable. The primary mRNA is subject to processing at the very end of the cggR open reading frame. This results in stable mature gapA and gapA-pgk-tpiA-pgm-eno mRNAs. PubMed The processing event requires the RNase Y PubMed.
- The intracellular concentration of CggR is about 230 nM (according to PubMed).
- The accumulation of the cggR-gapA mRNA is strongly dependent on the presence of the YkzW peptide, due to stabilization of the mRNA PubMed.
Biological materials
- Mutant:
- GP311 (in frame deletion), available in Stülke lab
- SM-NB7 (cggR-spc), available in Anne Galinier's and Boris Görke's labs
- GFP fusion:
- Antibody: available in Stülke lab
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References
Reviews
Original Publications
Matthias Gimpel, Nadja Heidrich, Ulrike Mäder, Hans Krügel, Sabine Brantl
A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon.
Mol Microbiol: 2010, 76(4);990-1009
[PubMed:20444087]
[WorldCat.org]
[DOI]
(I p)
Cédric Atmanene, Denix Chaix, Yannick Bessin, Nathalie Declerck, Alain Van Dorsselaer, Sarah Sanglier-Cianferani
Combination of noncovalent mass spectrometry and traveling wave ion mobility spectrometry reveals sugar-induced conformational changes of central glycolytic genes repressor/DNA complex.
Anal Chem: 2010, 82(9);3597-605
[PubMed:20361740]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Pavlína Rezácová, Milan Kozísek, Shiu F Moy, Irena Sieglová, Andrzej Joachimiak, Mischa Machius, Zbyszek Otwinowski
Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol Microbiol: 2008, 69(4);895-910
[PubMed:18554327]
[WorldCat.org]
[DOI]
(I p)
Thierry Doan, Laetitia Martin, Silvia Zorrilla, Denis Chaix, Stéphane Aymerich, Gilles Labesse, Nathalie Declerck
A phospho-sugar binding domain homologous to NagB enzymes regulates the activity of the central glycolytic genes repressor.
Proteins: 2008, 71(4);2038-50
[PubMed:18186488]
[WorldCat.org]
[DOI]
(I p)
Silvia Zorrilla, Denis Chaix, Alvaro Ortega, Carlos Alfonso, Thierry Doan, Emmanuel Margeat, Germán Rivas, Stephan Aymerich, Nathalie Declerck, Catherine A Royer
Fructose-1,6-bisphosphate acts both as an inducer and as a structural cofactor of the central glycolytic genes repressor (CggR).
Biochemistry: 2007, 46(51);14996-5008
[PubMed:18052209]
[WorldCat.org]
[DOI]
(P p)
Silvia Zorrilla, Thierry Doan, Carlos Alfonso, Emmanuel Margeat, Alvaro Ortega, Germán Rivas, Stéphane Aymerich, Catherine A Royer, Nathalie Declerck
Inducer-modulated cooperative binding of the tetrameric CggR repressor to operator DNA.
Biophys J: 2007, 92(9);3215-27
[PubMed:17293407]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Christoph Meinken, Hans-Matti Blencke, Holger Ludwig, Jörg Stülke
Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon.
Microbiology (Reading): 2003, 149(Pt 3);751-761
[PubMed:12634343]
[WorldCat.org]
[DOI]
(P p)
Thierry Doan, Stéphane Aymerich
Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate.
Mol Microbiol: 2003, 47(6);1709-21
[PubMed:12622823]
[WorldCat.org]
[DOI]
(P p)
Holger Ludwig, Nicole Rebhan, Hans-Matti Blencke, Matthias Merzbacher, Jörg Stülke
Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation.
Mol Microbiol: 2002, 45(2);543-53
[PubMed:12123463]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
S Fillinger, S Boschi-Muller, S Azza, E Dervyn, G Branlant, S Aymerich
Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium.
J Biol Chem: 2000, 275(19);14031-7
[PubMed:10799476]
[WorldCat.org]
[DOI]
(P p)
Additional publications: PubMed