Difference between revisions of "SpoIVCB"
(→References) |
|||
| Line 134: | Line 134: | ||
=References= | =References= | ||
| − | + | ==Reviews== | |
| + | <pubmed>20833318 </pubmed> | ||
| + | ==Original Publications== | ||
<pubmed>14526016,17720779,1900494,10438769,2492118,16385044,1691789,15383836,2536191,2163341,10931284,1577688,9852018,1942049,7814326,8550479,11959848,9501233,10400595,2115401,7592342,1518043,8226681,9078383,2514119,10611287,14523132, 19805276 10075739 7966271 17890309 </pubmed> | <pubmed>14526016,17720779,1900494,10438769,2492118,16385044,1691789,15383836,2536191,2163341,10931284,1577688,9852018,1942049,7814326,8550479,11959848,9501233,10400595,2115401,7592342,1518043,8226681,9078383,2514119,10611287,14523132, 19805276 10075739 7966271 17890309 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:06, 15 September 2010
- Description: sporulation-specific sigma factor (SigK) (N-terminal half), other part: spoIIIC
| Gene name | spoIVCB |
| Synonyms | cisB, sigK |
| Essential | no |
| Product | RNA polymerase sporulation-specific sigma factor (sigma-K) (N-terminal half) |
| Function | late mother cell-specific gene expression |
| MW, pI | 17 kDa, 8.208 |
| Gene length, protein length | 468 bp, 156 aa |
| Immediate neighbours | nucB, spoIVCA |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 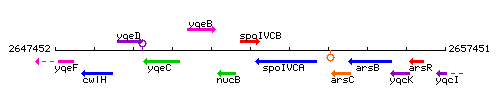
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU25760
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Genes controlled by SigK
gerE, lipC, surC, yabG, yndL, yitB-yitA-yisZ
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P12254
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Arthur Aronson, Purdue University, West Lafayette, USA homepage
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
Original Publications