Difference between revisions of "MreB"
(→Loclization) |
|||
| Line 129: | Line 129: | ||
==Reviews== | ==Reviews== | ||
<pubmed>17506674 17158703 15037301 </pubmed> | <pubmed>17506674 17158703 15037301 </pubmed> | ||
| − | == | + | ==Localization== |
<pubmed> 17064365, 20566861 </pubmed> | <pubmed> 17064365, 20566861 </pubmed> | ||
| + | |||
==Other original publications== | ==Other original publications== | ||
<pubmed>15752190,11290328,12809607,1400224,19192185,7836311, 16950129, 19192185, 19117023 19643765 19654094 19659933 8459776 11544518 20133608</pubmed> | <pubmed>15752190,11290328,12809607,1400224,19192185,7836311, 16950129, 19192185, 19117023 19643765 19654094 19659933 8459776 11544518 20133608</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:01, 19 July 2010
- Description: cell-shape determining protein
| Gene name | mreB |
| Synonyms | divIVB |
| Essential | yes PubMed |
| Product | cell-shape determining protein |
| Function | cell-shape determination |
| MW, pI | 35 kDa, 4.901 |
| Gene length, protein length | 1011 bp, 337 aa |
| Immediate neighbours | mreC, radC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 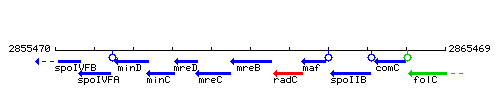
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28030
Phenotypes of a mutant
essential PubMed, the mutation can be suppressed by inactivation of ponA, ptsI, ccpA PubMed or addition of 5 mM Magnesium to the growth medium PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ftsA/mreB family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- UniProt: Q01465
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in the labs of Jeff Errington and Boris Görke
- Antibody: available in the Jeff Errington and Peter Graumann labs
Labs working on this gene/protein
Jeff Errington, Newcastle University, UK homepage
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Reviews
Localization
Other original publications
Hervé Joël Defeu Soufo, Christian Reimold, Uwe Linne, Tobias Knust, Johannes Gescher, Peter L Graumann
Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein.
Proc Natl Acad Sci U S A: 2010, 107(7);3163-8
[PubMed:20133608]
[WorldCat.org]
[DOI]
(I p)
Yoshikazu Kawai, Kei Asai, Jeffery Errington
Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis.
Mol Microbiol: 2009, 73(4);719-31
[PubMed:19659933]
[WorldCat.org]
[DOI]
(I p)
Daniel Muñoz-Espín, Richard Daniel, Yoshikazu Kawai, Rut Carballido-López, Virginia Castilla-Llorente, Jeff Errington, Wilfried J J Meijer, Margarita Salas
The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria.
Proc Natl Acad Sci U S A: 2009, 106(32);13347-52
[PubMed:19654094]
[WorldCat.org]
[DOI]
(I p)
Kathrin Schirner, Jeff Errington
Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 11);3611-3621
[PubMed:19643765]
[WorldCat.org]
[DOI]
(I p)
Yoshikazu Kawai, Richard A Daniel, Jeffery Errington
Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix.
Mol Microbiol: 2009, 71(5);1131-44
[PubMed:19192185]
[WorldCat.org]
[DOI]
(I p)
Joshua A Mayer, Kurt J Amann
Assembly properties of the Bacillus subtilis actin, MreB.
Cell Motil Cytoskeleton: 2009, 66(2);109-18
[PubMed:19117023]
[WorldCat.org]
[DOI]
(I p)
Rut Carballido-López, Alex Formstone, Ying Li, S Dusko Ehrlich, Philippe Noirot, Jeff Errington
Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE.
Dev Cell: 2006, 11(3);399-409
[PubMed:16950129]
[WorldCat.org]
[DOI]
(P p)
Alex Formstone, Jeffery Errington
A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis.
Mol Microbiol: 2005, 55(6);1646-57
[PubMed:15752190]
[WorldCat.org]
[DOI]
(P p)
Richard A Daniel, Jeff Errington
Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell.
Cell: 2003, 113(6);767-76
[PubMed:12809607]
[WorldCat.org]
[DOI]
(P p)
F van den Ent, L A Amos, J Löwe
Prokaryotic origin of the actin cytoskeleton.
Nature: 2001, 413(6851);39-44
[PubMed:11544518]
[WorldCat.org]
[DOI]
(P p)
L J Jones, R Carballido-López, J Errington
Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis.
Cell: 2001, 104(6);913-22
[PubMed:11290328]
[WorldCat.org]
[DOI]
(P p)
Y Abhayawardhane, G C Stewart
Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene.
J Bacteriol: 1995, 177(3);765-73
[PubMed:7836311]
[WorldCat.org]
[DOI]
(P p)
S Lee, C W Price
The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division.
Mol Microbiol: 1993, 7(4);601-10
[PubMed:8459776]
[WorldCat.org]
[DOI]
(P p)
P A Levin, P S Margolis, P Setlow, R Losick, D Sun
Identification of Bacillus subtilis genes for septum placement and shape determination.
J Bacteriol: 1992, 174(21);6717-28
[PubMed:1400224]
[WorldCat.org]
[DOI]
(P p)