Difference between revisions of "EpsC"
(→References) |
(→Other publications) |
||
| Line 126: | Line 126: | ||
==The EAR [[RNA switch]]== | ==The EAR [[RNA switch]]== | ||
<pubmed>20374491 20230605 </pubmed> | <pubmed>20374491 20230605 </pubmed> | ||
| − | == | + | ==Regulation of the ''eps'' operon== |
<pubmed>15661000,16430695,18047568,18647168 </pubmed> | <pubmed>15661000,16430695,18047568,18647168 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:02, 9 April 2010
- Description: extracellular polysaccharide synthesis
| Gene name | epsC |
| Synonyms | yveM |
| Essential | no |
| Product | unknown |
| Function | extracellular polysaccharide synthesis |
| Regulation of this protein in SubtiPathways: Biofilm | |
| MW, pI | 66 kDa, 8.775 |
| Gene length, protein length | 1794 bp, 598 aa |
| Immediate neighbours | epsD, epsB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 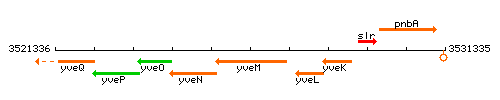
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU34350
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: polysaccharide synthase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P71052
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Richard Losick, Harvard Univ., Cambridge, USA homepage
Your additional remarks
References
The EAR RNA switch
Irnov Irnov, Wade C Winkler
A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales.
Mol Microbiol: 2010, 76(3);559-75
[PubMed:20374491]
[WorldCat.org]
[DOI]
(I p)
Zasha Weinberg, Joy X Wang, Jarrod Bogue, Jingying Yang, Keith Corbino, Ryan H Moy, Ronald R Breaker
Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes.
Genome Biol: 2010, 11(3);R31
[PubMed:20230605]
[WorldCat.org]
[DOI]
(I p)
Regulation of the eps operon