Difference between revisions of "IolF"
(→Expression and regulation) |
(→Expression and regulation) |
||
| Line 91: | Line 91: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' ''[[iolA]]-[[iolB]]-[[iolC]]-[[iolD]]-[[iolE]]-[[iolF]]-[[iolG]]-[[iolH]]-[[iolI]]-[[iolJ]]'' {{PubMed|9226270}} | ||
| − | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|9226270}} | |
| − | |||
| − | * '''[[Sigma factor]]:''' [[SigA]] | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** repressed by glucose ([[CcpA]]) | + | ** repressed by glucose ([[CcpA]]) {{PubMed|10666464}} |
** induced by inositol ([[IolR]]) {{PubMed|9887260,9226270}} | ** induced by inositol ([[IolR]]) {{PubMed|9887260,9226270}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
** [[IolR]]: transcription repression {{PubMed|9887260,9226270}} | ** [[IolR]]: transcription repression {{PubMed|9887260,9226270}} | ||
| − | ** [[CcpA]]: transcription repression | + | ** [[CcpA]]: transcription repression {{PubMed|10666464}} |
* '''Additional information:''' | * '''Additional information:''' | ||
Revision as of 22:17, 11 December 2009
- Description: inositol transport protein
| Gene name | iolF |
| Synonyms | yxdF |
| Essential | no |
| Product | inositol transport protein |
| Function | myo-inositol uptake |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 47 kDa, 9.474 |
| Gene length, protein length | 1317 bp, 439 aa |
| Immediate neighbours | iolG, iolE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 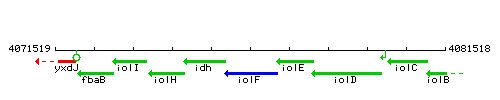
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU39710
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P42417
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Yasutaro Fujita, University of Fukuyama, Japan
- Ken-ichi Yoshida, Kobe University, Japan
Your additional remarks
References
Ken-ichi Yoshida, Masanori Yamaguchi, Tetsuro Morinaga, Masaki Kinehara, Maya Ikeuchi, Hitoshi Ashida, Yasutaro Fujita
myo-Inositol catabolism in Bacillus subtilis.
J Biol Chem: 2008, 283(16);10415-24
[PubMed:18310071]
[WorldCat.org]
[DOI]
(P p)
Ken-Ichi Yoshida, Yoshiyuki Yamamoto, Kaoru Omae, Mami Yamamoto, Yasutaro Fujita
Identification of two myo-inositol transporter genes of Bacillus subtilis.
J Bacteriol: 2002, 184(4);983-91
[PubMed:11807058]
[WorldCat.org]
[DOI]
(P p)
K I Yoshida, T Shibayama, D Aoyama, Y Fujita
Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon.
J Mol Biol: 1999, 285(3);917-29
[PubMed:9887260]
[WorldCat.org]
[DOI]
(P p)
K I Yoshida, D Aoyama, I Ishio, T Shibayama, Y Fujita
Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis.
J Bacteriol: 1997, 179(14);4591-8
[PubMed:9226270]
[WorldCat.org]
[DOI]
(P p)