Difference between revisions of "DivIVA"
(→Additional information) |
(→The protein) |
||
| Line 53: | Line 53: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' DivIVA is required for polar localisation of MinCD via MinJ. It also recruits RacA to the distal pole of the prespore. | + | * '''Catalyzed reaction/ biological activity:''' DivIVA is required for polar localisation of MinCD via MinJ. [http://www.ncbi.nlm.nih.gov/sites/entrez/19019154 PubMed] It also recruits RacA to the distal pole of the prespore [http://www.ncbi.nlm.nih.gov/sites/entrez/12493822 PubMed]. |
* '''Protein family:''' gpsB family (according to Swiss-Prot) | * '''Protein family:''' gpsB family (according to Swiss-Prot) | ||
| Line 64: | Line 64: | ||
* '''Domains:''' The first 60 amino acids constitute a lipid binding domain. [http://www.ncbi.nlm.nih.gov/sites/entrez/19478798 PubMed] | * '''Domains:''' The first 60 amino acids constitute a lipid binding domain. [http://www.ncbi.nlm.nih.gov/sites/entrez/19478798 PubMed] | ||
| − | Multimerisation involves two coiled-coil motifs, one in the lipid binding domain, and the other one being present in the helical C-terminal domain. | + | Multimerisation involves two coiled-coil motifs, one in the lipid binding domain, and the other one being present in the helical C-terminal domain [http://www.ncbi.nlm.nih.gov/sites/entrez/18388125 PubMed]. |
| − | * '''Modification:''' The Mycobacterium DivIVA homologue Wag31 is phosphorylated at T73. | + | * '''Modification:''' The Mycobacterium DivIVA homologue Wag31 is phosphorylated at T73 [http://www.ncbi.nlm.nih.gov/sites/entrez/15985609 PubMed]. |
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' not known |
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' not known |
* '''Interactions:''' [[DivIVA]]-[[RacA]], [[DivIVA]]-[[MinJ]], [[DivIVA]]-[[DivIVA]] | * '''Interactions:''' [[DivIVA]]-[[RacA]], [[DivIVA]]-[[MinJ]], [[DivIVA]]-[[DivIVA]] | ||
| − | * '''Localization:''' DivIVA forms a ring underneath the invaginating membrane at the site of cell division and is enriched at both cell poles. | + | * '''Localization:''' DivIVA forms a ring underneath the invaginating membrane at the site of cell division and is enriched at both cell poles [http://www.ncbi.nlm.nih.gov/sites/entrez/9219999 PubMed]. |
=== Database entries === | === Database entries === | ||
Revision as of 08:37, 27 August 2009
- Description: cell-division initiation protein (septum placement)
| Gene name | divIVA |
| Synonyms | ylmJ |
| Essential | no |
| Product | cell-division initiation protein |
| Function | septum placement |
| MW, pI | 19 kDa, 4.846 |
| Gene length, protein length | 492 bp, 164 aa |
| Immediate neighbours | ylmH, ileS |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 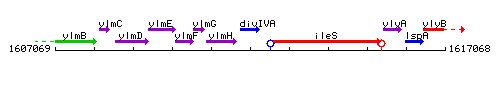
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU15420
Phenotypes of a mutant
Deletion of divIVA leads to filamentation and polar divisions that in turn cause a minicell phenotype. A divIVA mutant has a severe sporulation defect.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
filamentation is suppressed by minCD mutations
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: DivIVA is required for polar localisation of MinCD via MinJ. PubMed It also recruits RacA to the distal pole of the prespore PubMed.
- Protein family: gpsB family (according to Swiss-Prot)
- Paralogous protein(s): GpsB
Extended information on the protein
- Kinetic information:
- Domains: The first 60 amino acids constitute a lipid binding domain. PubMed
Multimerisation involves two coiled-coil motifs, one in the lipid binding domain, and the other one being present in the helical C-terminal domain PubMed.
- Modification: The Mycobacterium DivIVA homologue Wag31 is phosphorylated at T73 PubMed.
- Cofactor(s): not known
- Effectors of protein activity: not known
- Localization: DivIVA forms a ring underneath the invaginating membrane at the site of cell division and is enriched at both cell poles PubMed.
Database entries
- Structure:
- UniProt: P71021
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: one gene cistron
- Additional information:
Biological materials
- Mutant: divIVA::tet available from the Hamoen Lab
- Expression vector:
- lacZ fusion:
- GFP fusion: divIVA-gfp fusions available from the Hamoen Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Centre for Bacterial Cell Biology, Newcastle upon Tyne, United Kingdom [4]
Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
Your additional remarks
References
Kumaran S Ramamurthi, Richard Losick
Negative membrane curvature as a cue for subcellular localization of a bacterial protein.
Proc Natl Acad Sci U S A: 2009, 106(32);13541-5
[PubMed:19666580]
[WorldCat.org]
[DOI]
(I p)
Jennifer R Juarez, William Margolin
Irresistible curves.
EMBO J: 2009, 28(15);2147-8
[PubMed:19654604]
[WorldCat.org]
[DOI]
(I p)
Rok Lenarcic, Sven Halbedel, Loek Visser, Michael Shaw, Ling Juan Wu, Jeff Errington, Davide Marenduzzo, Leendert W Hamoen
Localisation of DivIVA by targeting to negatively curved membranes.
EMBO J: 2009, 28(15);2272-82
[PubMed:19478798]
[WorldCat.org]
[DOI]
(I p)
Pamela Gamba, Jan-Willem Veening, Nigel J Saunders, Leendert W Hamoen, Richard A Daniel
Two-step assembly dynamics of the Bacillus subtilis divisome.
J Bacteriol: 2009, 191(13);4186-94
[PubMed:19429628]
[WorldCat.org]
[DOI]
(I p)
Marc Bramkamp, Robyn Emmins, Louise Weston, Catriona Donovan, Richard A Daniel, Jeff Errington
A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD.
Mol Microbiol: 2008, 70(6);1556-69
[PubMed:19019154]
[WorldCat.org]
[DOI]
(I p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Leendert W Hamoen, Jeffery Errington
Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B.
J Bacteriol: 2003, 185(2);693-7
[PubMed:12511520]
[WorldCat.org]
[DOI]
(P p)
Frederico J Gueiros-Filho, Richard Losick
A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ.
Genes Dev: 2002, 16(19);2544-56
[PubMed:12368265]
[WorldCat.org]
[DOI]
(P p)
H B Thomaides, M Freeman, M El Karoui, J Errington
Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation.
Genes Dev: 2001, 15(13);1662-73
[PubMed:11445541]
[WorldCat.org]
[DOI]
(P p)
D H Edwards, H B Thomaides, J Errington
Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast.
EMBO J: 2000, 19(11);2719-27
[PubMed:10835369]
[WorldCat.org]
[DOI]
(P p)
D H Edwards, J Errington
The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division.
Mol Microbiol: 1997, 24(5);905-15
[PubMed:9219999]
[WorldCat.org]
[DOI]
(P p)