Difference between revisions of "MinC"
(→References) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' cell-division inhibitor (septum placement), destabilizes FtsZ-rings at new cell poles <br/><br/> | + | * '''Description:''' cell-division inhibitor (septum placement), destabilizes [[FtsZ]]-rings at new cell poles <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 119: | Line 119: | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed> 19680248 </pubmed> | + | <pubmed> 19680248 19884039 </pubmed> |
| − | |||
==Original Publications== | ==Original Publications== | ||
<pubmed>8459776,19019154,1400224,10411726,15317782,,18588879, 19141479 </pubmed> | <pubmed>8459776,19019154,1400224,10411726,15317782,,18588879, 19141479 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:25, 4 November 2009
- Description: cell-division inhibitor (septum placement), destabilizes FtsZ-rings at new cell poles
| Gene name | minC |
| Synonyms | |
| Essential | no |
| Product | cell-division inhibitor |
| Function | septum placement |
| MW, pI | 24 kDa, 6.262 |
| Gene length, protein length | 678 bp, 226 aa |
| Immediate neighbours | minD, mreD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 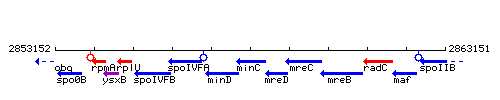
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28000
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: minC family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: Q01463
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications