Difference between revisions of "CggR"
| Line 81: | Line 81: | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2OKG 2OKG] ( effector binding domain), [http://www.rcsb.org/pdb/explore.do?structureId=3BXH 3BXH] (in complex with fructose-6-phosphate), complex with Fructose-6-Phosphate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=65242 NCBI], effector binding domain [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=44226 NCBI] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2OKG 2OKG] ( effector binding domain), [http://www.rcsb.org/pdb/explore.do?structureId=3BXH 3BXH] (in complex with fructose-6-phosphate), complex with Fructose-6-Phosphate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=65242 NCBI], effector binding domain [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=44226 NCBI] | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/O32253 O32253] |
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU33950] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU33950] | ||
Revision as of 14:35, 20 July 2009
- Description: repressor of the glycolytic gapA operon
| Gene name | cggR |
| Synonyms | yvbQ |
| Essential | no |
| Product | central glycolytic genes regulator |
| Function | transcriptional regulator |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 37,2 kDa,5.68 |
| Gene length, protein length | 1020 bp, 340 amino acids |
| Immediate neighbours | araE, gapA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 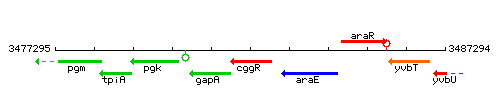
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU33950
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription repression of the glycolytic gapA operon
- Protein family: sorC transcriptional regulatory family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- DNA binding domain (H-T-H motif) (37–56)
- Modification:
- Cofactor(s):
- Effectors of protein activity: fructose 1.6-bisphosphate PubMed and dihydroxyacetone phosphate, glucose-6-phosphate and fructose-6-phosphate PubMed act as inducer and result in release of CggR from the DNA
- Interactions:
- Localization:
Database entries
- Structure: 2OKG ( effector binding domain), 3BXH (in complex with fructose-6-phosphate), complex with Fructose-6-Phosphate NCBI, effector binding domain NCBI
- UniProt: O32253
- KEGG entry: [3]
Additional information
Expression and regulation
The primary mRNAs of the operon are highly unstable. The primary mRNA is subject to processing at the very end of the cggR open reading frame. This results in stable mature gapA and gapA-pgk-tpiA-pgm-eno mRNAs. The processing event requires the Rny protein.
- Sigma factor: SigA
- Regulation: expression activated by glucose (77 fold) PubMed, CggR represses the operon in the absence of glycolytic sugars PubMed
- Regulatory mechanism: repression
- Database entries: DBTBS
- Additional information:
Biological materials
- Mutant: GP311 (in frame deletion), available in Stülke lab
- GFP fusion:
- Antibody: available in Stülke lab
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References