Difference between revisions of "OhrR"
(→References) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 147: | Line 147: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed>22797754 20626317 19575568 20094649 </pubmed> | + | <pubmed>22797754 20626317 19575568 20094649 25852656</pubmed> |
==Original Publications== | ==Original Publications== | ||
<pubmed>21749987,16209951,18487332,17660290,17502599,12486061,11418552,18586944,11983871,18363800,19129220 9696771 21749987 22797754 24313874 </pubmed> | <pubmed>21749987,16209951,18487332,17660290,17502599,12486061,11418552,18586944,11983871,18363800,19129220 9696771 21749987 22797754 24313874 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 11:02, 10 April 2015
- Description: transcription repressor of the ohrA gene
| Gene name | ohrR |
| Synonyms | ykmA |
| Essential | no |
| Product | transcription repressor (MarR family) |
| Function | regulation of ohrA expression in response to organic peroxides |
| Gene expression levels in SubtiExpress: ohrR | |
| Interactions involving this protein in SubtInteract: OhrR | |
| MW, pI | 16 kDa, 6.364 |
| Gene length, protein length | 441 bp, 147 aa |
| Immediate neighbours | ohrA, ohrB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 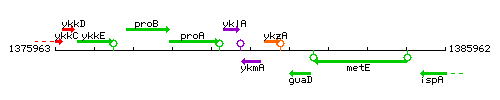
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed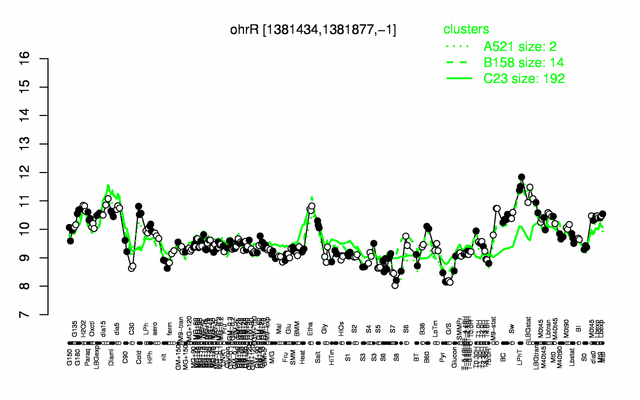
| |
Contents
Categories containing this gene/protein
transcription factors and their control, resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The OhrR regulon:
The gene
Basic information
- Locus tag: BSU13150
Phenotypes of a mutant
Database entries
- BsubCyc: BSU13150
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MarR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU13150
- UniProt: O34777
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: ohrR PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 133 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Richard Brennan, Houston, Texas, USA Homepage
Your additional remarks
References
Reviews
Vu Van Loi, Martina Rossius, Haike Antelmann
Redox regulation by reversible protein S-thiolation in bacteria.
Front Microbiol: 2015, 6;187
[PubMed:25852656]
[WorldCat.org]
[DOI]
(P e)
James M Dubbs, Skorn Mongkolsuk
Peroxide-sensing transcriptional regulators in bacteria.
J Bacteriol: 2012, 194(20);5495-503
[PubMed:22797754]
[WorldCat.org]
[DOI]
(I p)
Haike Antelmann, John D Helmann
Thiol-based redox switches and gene regulation.
Antioxid Redox Signal: 2011, 14(6);1049-63
[PubMed:20626317]
[WorldCat.org]
[DOI]
(I p)
Victor Duarte, Jean-Marc Latour
PerR vs OhrR: selective peroxide sensing in Bacillus subtilis.
Mol Biosyst: 2010, 6(2);316-23
[PubMed:20094649]
[WorldCat.org]
[DOI]
(I p)
Peter Zuber
Management of oxidative stress in Bacillus.
Annu Rev Microbiol: 2009, 63;575-97
[PubMed:19575568]
[WorldCat.org]
[DOI]
(I p)
Original Publications
Ahmed Gaballa, Bui Khanh Chi, Alexandra A Roberts, Dörte Becher, Chris J Hamilton, Haike Antelmann, John D Helmann
Redox regulation in Bacillus subtilis: The bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE.
Antioxid Redox Signal: 2014, 21(3);357-67
[PubMed:24313874]
[WorldCat.org]
[DOI]
(I p)
James M Dubbs, Skorn Mongkolsuk
Peroxide-sensing transcriptional regulators in bacteria.
J Bacteriol: 2012, 194(20);5495-503
[PubMed:22797754]
[WorldCat.org]
[DOI]
(I p)
Bui Khanh Chi, Katrin Gronau, Ulrike Mäder, Bernd Hessling, Dörte Becher, Haike Antelmann
S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics.
Mol Cell Proteomics: 2011, 10(11);M111.009506
[PubMed:21749987]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, Sumarin Soonsanga, Jin-Won Lee, John D Helmann
Oxidation of a single active site suffices for the functional inactivation of the dimeric Bacillus subtilis OhrR repressor in vitro.
Nucleic Acids Res: 2009, 37(4);1174-81
[PubMed:19129220]
[WorldCat.org]
[DOI]
(I p)
Sumarin Soonsanga, Jin-Won Lee, John D Helmann
Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor.
J Bacteriol: 2008, 190(17);5738-45
[PubMed:18586944]
[WorldCat.org]
[DOI]
(I p)
Falko Hochgräfe, Carmen Wolf, Stephan Fuchs, Manuel Liebeke, Michael Lalk, Susanne Engelmann, Michael Hecker
Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus.
J Bacteriol: 2008, 190(14);4997-5008
[PubMed:18487332]
[WorldCat.org]
[DOI]
(I p)
Sumarin Soonsanga, Jin-Won Lee, John D Helmann
Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR.
Mol Microbiol: 2008, 68(4);978-86
[PubMed:18363800]
[WorldCat.org]
[DOI]
(I p)
Sumarin Soonsanga, Mayuree Fuangthong, John D Helmann
Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR.
J Bacteriol: 2007, 189(19);7069-76
[PubMed:17660290]
[WorldCat.org]
[DOI]
(P p)
Jin-Won Lee, Sumarin Soonsanga, John D Helmann
A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR.
Proc Natl Acad Sci U S A: 2007, 104(21);8743-8
[PubMed:17502599]
[WorldCat.org]
[DOI]
(P p)
Minsun Hong, Mayuree Fuangthong, John D Helmann, Richard G Brennan
Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family.
Mol Cell: 2005, 20(1);131-41
[PubMed:16209951]
[WorldCat.org]
[DOI]
(P p)
John D Helmann, Ming Fang Winston Wu, Ahmed Gaballa, Phil A Kobel, Maud M Morshedi, Paul Fawcett, Chris Paddon
The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors.
J Bacteriol: 2003, 185(1);243-53
[PubMed:12486061]
[WorldCat.org]
[DOI]
(P p)
Mayuree Fuangthong, John D Helmann
The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative.
Proc Natl Acad Sci U S A: 2002, 99(10);6690-5
[PubMed:11983871]
[WorldCat.org]
[DOI]
(P p)
M Fuangthong, S Atichartpongkul, S Mongkolsuk, J D Helmann
OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis.
J Bacteriol: 2001, 183(14);4134-41
[PubMed:11418552]
[WorldCat.org]
[DOI]
(P p)
U Völker, K K Andersen, H Antelmann, K M Devine, M Hecker
One of two osmC homologs in Bacillus subtilis is part of the sigmaB-dependent general stress regulon.
J Bacteriol: 1998, 180(16);4212-8
[PubMed:9696771]
[WorldCat.org]
[DOI]
(P p)