Difference between revisions of "Rnz"
(→Original Publications) |
|||
| (17 intermediate revisions by 3 users not shown) | |||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || processing of CCA-less tRNA precursors | |style="background:#ABCDEF;" align="center"|'''Function''' || processing of CCA-less tRNA precursors | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23840 rnz] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 33 kDa, 5.805 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 33 kDa, 5.805 | ||
| Line 18: | Line 20: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 921 bp, 307 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 921 bp, 307 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rpmGC]]'', ''[[zwf]]'' |
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU23840 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU23840 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU23840 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yqjK_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yqjK_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnz_2478006_2478929_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:rnz_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23840]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/><br/><br/> | |
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
<br/><br/> | <br/><br/> | ||
| Line 50: | Line 56: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU23840&redirect=T BSU23840] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqjK.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqjK.html] | ||
| Line 56: | Line 63: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 64: | Line 68: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' Endonucleolytic cleavage of RNA, removing extra 3' nucleotides from tRNA precursor, generating 3' termini of tRNAs (according to Swiss-Prot) | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** Endonucleolytic cleavage of RNA, removing extra 3' nucleotides from tRNA precursor, generating 3' termini of tRNAs (according to Swiss-Prot) | ||
| + | ** 3' end maturation of tmRNA {{PubMed|25402410}} | ||
* '''Protein family:''' [[RNase]] Z family (according to Swiss-Prot) | * '''Protein family:''' [[RNase]] Z family (according to Swiss-Prot) | ||
| Line 82: | Line 88: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU23840&redirect=T BSU23840] | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2FK6 2FK6] (in complex with tRNA-Thr), [http://www.rcsb.org/pdb/explore.do?structureId=1Y44 1Y44] | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2FK6 2FK6] (in complex with tRNA-Thr) {{PubMed|22940585}}, [http://www.rcsb.org/pdb/explore.do?structureId=1Y44 1Y44] |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P54548 P54548] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P54548 P54548] | ||
| Line 102: | Line 109: | ||
* '''Operon:''' ''rnz'' (according to [http://dbtbs.hgc.jp/COG/prom/yqjK.html DBTBS]) | * '''Operon:''' ''rnz'' (according to [http://dbtbs.hgc.jp/COG/prom/yqjK.html DBTBS]) | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnz_2478006_2478929_-1 rnz] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 109: | Line 118: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 55 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 278 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 379 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 311 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
| Line 132: | Line 144: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed> 19215776 17599240 </pubmed> | + | <pubmed> 19215776 17599240 25878039</pubmed> |
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>17005971 12941704 15654328 16518398 </pubmed> | + | <pubmed>17005971 12941704 15654328 16518398 22940585 25402410 24022488</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 15:09, 26 April 2015
- Description: RNase Z
| Gene name | rnz |
| Synonyms | yqjK |
| Essential | yes PubMed |
| Product | endoribonuclease Z |
| Function | processing of CCA-less tRNA precursors |
| Gene expression levels in SubtiExpress: rnz | |
| MW, pI | 33 kDa, 5.805 |
| Gene length, protein length | 921 bp, 307 aa |
| Immediate neighbours | rpmGC, zwf |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed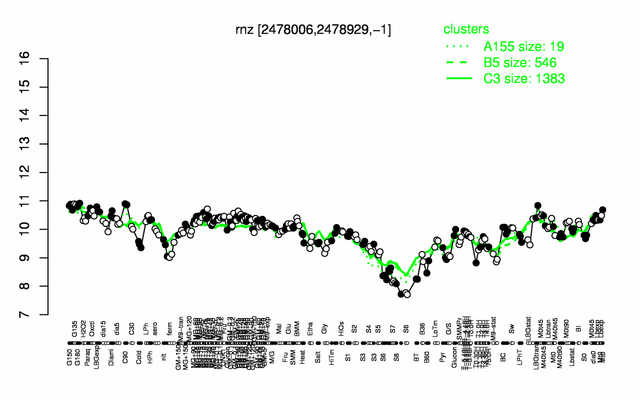
| |
Contents
Categories containing this gene/protein
Rnases, translation, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23840
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU23840
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Endonucleolytic cleavage of RNA, removing extra 3' nucleotides from tRNA precursor, generating 3' termini of tRNAs (according to Swiss-Prot)
- 3' end maturation of tmRNA PubMed
- Protein family: RNase Z family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU23840
- UniProt: P54548
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: rnz (according to DBTBS)
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 55 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 278 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 379 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 311 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Ciaran Condon, IBPC, Paris, France Homepage
Your additional remarks
References
Reviews
Murray P Deutscher
How bacterial cells keep ribonucleases under control.
FEMS Microbiol Rev: 2015, 39(3);350-61
[PubMed:25878039]
[WorldCat.org]
[DOI]
(I p)
Roland K Hartmann, Markus Gössringer, Bettina Späth, Susan Fischer, Anita Marchfelder
The making of tRNAs and more - RNase P and tRNase Z.
Prog Mol Biol Transl Sci: 2009, 85;319-68
[PubMed:19215776]
[WorldCat.org]
[DOI]
(P p)
B Späth, G Canino, A Marchfelder
tRNase Z: the end is not in sight.
Cell Mol Life Sci: 2007, 64(18);2404-12
[PubMed:17599240]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Laetitia Gilet, Jeanne M DiChiara, Sabine Figaro, David H Bechhofer, Ciarán Condon
Small stable RNA maturation and turnover in Bacillus subtilis.
Mol Microbiol: 2015, 95(2);270-82
[PubMed:25402410]
[WorldCat.org]
[DOI]
(I p)
Tanmay Dutta, Arun Malhotra, Murray P Deutscher
How a CCA sequence protects mature tRNAs and tRNA precursors from action of the processing enzyme RNase BN/RNase Z.
J Biol Chem: 2013, 288(42);30636-30644
[PubMed:24022488]
[WorldCat.org]
[DOI]
(I p)
Olivier Pellegrini, Inés Li de la Sierra-Gallay, Jérémie Piton, Laetitia Gilet, Ciarán Condon
Activation of tRNA maturation by downstream uracil residues in B. subtilis.
Structure: 2012, 20(10);1769-77
[PubMed:22940585]
[WorldCat.org]
[DOI]
(I p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)
Inés Li de la Sierra-Gallay, Nathalie Mathy, Olivier Pellegrini, Ciarán Condon
Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat Struct Mol Biol: 2006, 13(4);376-7
[PubMed:16518398]
[WorldCat.org]
[DOI]
(P p)
Inés Li de la Sierra-Gallay, Olivier Pellegrini, Ciarán Condon
Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Nature: 2005, 433(7026);657-61
[PubMed:15654328]
[WorldCat.org]
[DOI]
(I p)
Olivier Pellegrini, Jamel Nezzar, Anita Marchfelder, Harald Putzer, Ciarán Condon
Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis.
EMBO J: 2003, 22(17);4534-43
[PubMed:12941704]
[WorldCat.org]
[DOI]
(P p)