Difference between revisions of "PrkC"
| Line 125: | Line 125: | ||
<pubmed>19246764,12842463,,12406230,16025310, 17218307, 12399479, 12406230, 19246764, 12842463 , 18984160 </pubmed> | <pubmed>19246764,12842463,,12406230,16025310, 17218307, 12399479, 12406230, 19246764, 12842463 , 18984160 </pubmed> | ||
| − | |||
| − | |||
Revision as of 17:52, 15 June 2009
- Description: protein kinase C
| Gene name | prkC |
| Synonyms | yloP |
| Essential | no |
| Product | protein kinase |
| Function | germination in response to muropeptides |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 71 kDa, 4.833 |
| Gene length, protein length | 1944 bp, 648 aa |
| Immediate neighbours | prpC, cpgA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 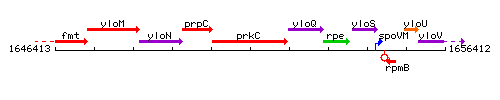
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU15770
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + a protein = ADP + a phosphoprotein (according to Swiss-Prot)
- Protein family: protein kinase domain (according to Swiss-Prot)
- Paralogous protein(s):
Proteins phosphorylated by PrkC
CpgA, EF-Tu, YezB PubMed, EF-G PubMed
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Thr-290 PubMed
- Cofactor(s):
- Effectors of protein activity: activated by muropeptides PubMed
- Interactions:
- Localization: membrane (according to Swiss-Prot), membrane PubMed
Database entries
- Structure:
- Swiss prot entry: O34507
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector: pGP832 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References