Difference between revisions of "SerA"
| Line 52: | Line 52: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' 3-phospho-D-glycerate + NAD<sup>+</sup> = 3-phosphonooxypyruvate + NADH (according to Swiss-Prot) |
* '''Protein family:''' D-isomer specific 2-hydroxyacid dehydrogenase family (according to Swiss-Prot) | * '''Protein family:''' D-isomer specific 2-hydroxyacid dehydrogenase family (according to Swiss-Prot) | ||
Revision as of 23:59, 23 May 2009
- Description: phosphoglycerate dehydrogenase
| Gene name | serA |
| Synonyms | |
| Essential | no |
| Product | phosphoglycerate dehydrogenase |
| Function | biosynthesis of serine |
| MW, pI | 56 kDa, 5.617 |
| Gene length, protein length | 1575 bp, 525 aa |
| Immediate neighbours | ypzE, aroC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 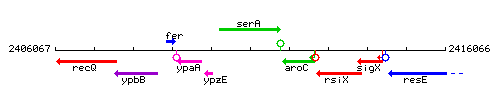
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 3-phospho-D-glycerate + NAD+ = 3-phosphonooxypyruvate + NADH (according to Swiss-Prot)
- Protein family: D-isomer specific 2-hydroxyacid dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: S-cysteinylation after diamide stress (C410)PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: membrane PubMed
Database entries
- Structure:
- Swiss prot entry: P35136
- KEGG entry: BSU23070
- E.C. number: 1.1.1.95
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: