Difference between revisions of "DnaX"
(→Extended information on the protein) |
|||
| Line 74: | Line 74: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' [[DnaX]]-[[DnaB]] [http://www.ncbi.nlm.nih.gov/sites/entrez/14757052 PubMed] |
* '''Localization:''' Nucleoid (Mid-cell) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] nucleoid region (mid-cell spot) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | * '''Localization:''' Nucleoid (Mid-cell) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] nucleoid region (mid-cell spot) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | ||
Revision as of 12:27, 28 April 2009
- Description: DNA polymerase III (gamma and tau subunits)
| Gene name | dnaX |
| Synonyms | dnaH, dna-8132 |
| Essential | yes PubMed |
| Product | DNA polymerase III (gamma and tau subunits) |
| Function | DNA replication |
| MW, pI | 62 kDa, 5.488 |
| Gene length, protein length | 1689 bp, 563 aa |
| Immediate neighbours | scr, yaaK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 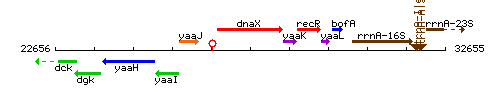
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Haroniti et al. (2004) The clamp-loader-helicase interaction in Bacillus. Atomic force microscopy reveals the structural organisation of the DnaB-tau complex in Bacillus.J. Mol. Biol. 336: 381-393. PubMed
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed