Difference between revisions of "Obg"
(→Database entries) |
(→Basic information/ Evolution) |
||
| Line 54: | Line 54: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' Binds and hydrolyzes GTP and readily exchanges GDP for GTP |
| − | Binds and hydrolyzes GTP and readily exchanges GDP for GTP | ||
| − | * '''Protein family:'''Era/Obg family | + | * '''Protein family:''' Era/Obg family |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[Era]], [[Obg]], [[YphC]], [[YsxC]], [[YlqF]], [[YqeH]], [[ThdF]], [[YyaF]], [[YnbA]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
Revision as of 14:48, 24 February 2009
- Description: GTP-binding protein involved in initiation of sporulation
| Gene name | obg |
| Synonyms | |
| Essential | Yes |
| Product | GTP-binding protein |
| Function | may be required to stimulate activity of the phosphorelay that activates Spo0A |
| MW, pI | 47 kDa, 4.859 |
| Gene length, protein length | 1284 bp, 428 aa |
| Immediate neighbours | spo0B, pheB |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 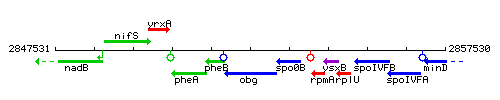
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Binds and hydrolyzes GTP and readily exchanges GDP for GTP
- Protein family: Era/Obg family
Extended information on the protein
- Kinetic information:
- Domains:
Obg possesses 3 domains: A N-terminal glycine-rich domain (1–158), a GTP binding domain (159–342) and a C-terminal TGS domain (343–428). The TGS domain has no known function, but it is named for three protein families in which it is found, ThrRS, GTPase, and SpoT.
- Modification:
- Cofactor(s):
GDP, GTP and probably ppGpp
- Effectors of protein activity:
- Interactions:
Interaction with Rsb proteins.
- Localization:
Database entries
- Structure:1LNZ
- Swiss prot entry:[3]
- KEGG entry:[4]
- E.C. number:
Additional information
The biochemical analysis of bacterial and human Obg proteins combined with the structural observation of the ppGpp nucleotide within the Obg active site suggest a potential role for ppGpp modulation of Obg function in B. subtilis.
Expression and regulation
- Sigma factor: SigA
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Buglino J., Shen V., Hakiman P. & Lima C.D. (2002) Structural and biochemical analysis of the Obg GTP binding protein Structure 10: 1581-1592. PubMed
- Scott, J.M., Ju, J., Mitchell, T., and Haldenwang, W.G. (2000) The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes J. Bacteriol. 182: 2771–2777. PubMed