Difference between revisions of "LevD"
| Line 18: | Line 18: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 438 bp, 146 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 438 bp, 146 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[levE]]'', ''[[levR]]'' |
|- | |- | ||
|style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/levD_nucleotide.txt Gene sequence (+200bp) ]''' | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/levD_nucleotide.txt Gene sequence (+200bp) ]''' | ||
Revision as of 16:32, 20 February 2009
- Description: write here
| Gene name | levD |
| Synonyms | sacL |
| Essential | no |
| Product | phosphotransferase system (PTS) fructose-specific enzyme IIA |
| Function | |
| MW, pI | 16 kDa, 4.479 |
| Gene length, protein length | 438 bp, 146 aa |
| Immediate neighbours | levE, levR |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 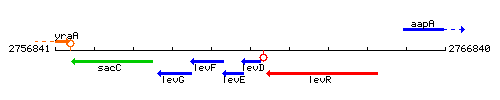
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry:
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Sigma factor: SigL
- Regulation: carbon catabolite repression, induction by fructose
- Regulatory mechanism: catabolite repression: transcription repression by CcpA, transcription activator LevR is less active in the presence of glucose; induction: transcription activation by LevR
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Martin-Verstraete, I., Débarbouillé, M., Klier, A., and Rapoport, G. (1990) Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol 214: 657-671. PubMed
- Martin-Verstraete, I. M. Débarbouillé, A. Klier, and G. Rapoport. 1992. Mutagenesis of the Bacillus subtilis ˝-12, -24˝ promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J. Mol. Biol. 226: 85-99. PubMed
- Stülke, J., Martin-Verstraete, I., Charrier, V., Klier, A., Deutscher, J. & Rapoport, G. (1995) The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177: 6928-6936. PubMed
- Martin-Verstraete, I., Stülke, J., Klier, A. & Rapoport, G. (1995) Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177: 6919-6927. PubMed
- Martin-Verstraete, I., Charrier, V., Stülke, J., Galinier, A., Erni, B., Rapoport, G., & Deutscher, J. (1998) Antagonistic effects of dual PTS catalyzed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol. Microbiol. 28: 293-303. PubMed
- Charrier V, Deutscher J, Martin-Verstraete I (1997b) Protein phosphorylation chain of a Bacillus subtilis fructose-specific phosphotransferase system and its participation in regulation of the expression of the lev operon. Biochemistry 36:1163-1172. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed