Difference between revisions of "KtrA"
(→Original publications) |
(→Original publications) |
||
| Line 157: | Line 157: | ||
<pubmed>25869574 </pubmed> | <pubmed>25869574 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>12562800,15096624,16321950, 20511502, 23086297 23598340 16990138 24141192 25957408</pubmed> | + | <pubmed>12562800,15096624,16321950, 20511502, 23086297 23598340 16990138 24141192 25957408 16990138</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:20, 14 May 2015
- Description: high affinity potassium transporter KtrA-KtrB, peripheric membrane component (proton symport)
| Gene name | ktrA |
| Synonyms | yuaA |
| Essential | no |
| Product | high affinity potassium transporter KtrA-KtrB, peripheric membrane component (proton symport) |
| Function | potassium uptake |
| Gene expression levels in SubtiExpress: ktrA | |
| Interactions involving this protein in SubtInteract: KtrA | |
| Metabolic function and regulation of this protein in SubtiPathways: Metal ion homeostasis, ktrA | |
| MW, pI | 24 kDa, 5.981 |
| Gene length, protein length | 666 bp, 222 aa |
| Immediate neighbours | bslA, ktrB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 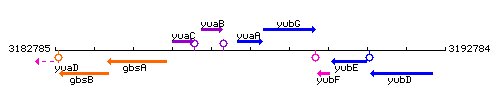
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed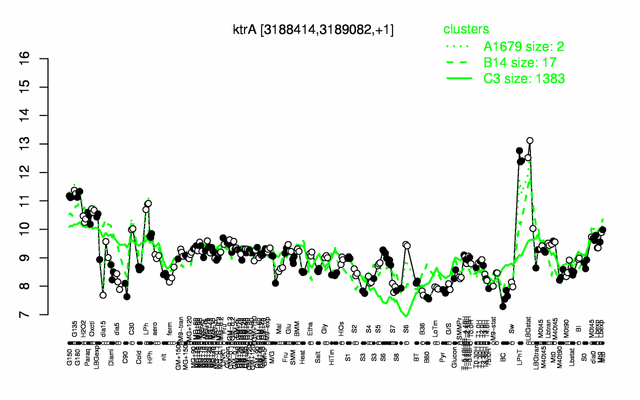
| |
Contents
Categories containing this gene/protein
transporters/ other, metal ion homeostasis (K, Na, Ca, Mg), coping with hyper-osmotic stress, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU31090
Phenotypes of a mutant
Database entries
- BsubCyc: BSU31090
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): KtrC
Extended information on the protein
- Kinetic information:
- Domains:
- contains a RCK_N domain at the N-terminus (according to UniProt, [2])
- contains a RCK_C domain at the C-terminus (according to UniProt, [3])
- Modification:
- Effectors of protein activity:
- Localization: peripheral membrane protein PubMed
Database entries
- BsubCyc: BSU31090
- UniProt: O32080
- KEGG entry: [4]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- expression is controlled via termination antitermination by the ydaO riboswitch, expression is switched off upon binding of c-di-AMP PubMed
- Additional information:
Biological materials
- Mutant:
- 1A954 ( ktrA::kan), PubMed, available at BGSC
- GHB1 (D(ktrA-ktrB)::aphA3), available in Erhard Bremer's lab
- GP92 (D(ktrA-ktrB)::aphA3), available in Jörg Stülke's lab
- Expression vector:
- pGP2594: (IPTG inducible expression, purification in E. coli with N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Erhard Bremer, University of Marburg, Germany homepage
Your additional remarks
References
Reviews
Fabian M Commichau, Achim Dickmanns, Jan Gundlach, Ralf Ficner, Jörg Stülke
A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP.
Mol Microbiol: 2015, 97(2);189-204
[PubMed:25869574]
[WorldCat.org]
[DOI]
(I p)
Original publications
Henna Kim, Suk-Jun Youn, Seong Ok Kim, Junsang Ko, Jie-Oh Lee, Byong-Seok Choi
Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C Domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP).
J Biol Chem: 2015, 290(26);16393-402
[PubMed:25957408]
[WorldCat.org]
[DOI]
(I p)
James W Nelson, Narasimhan Sudarsan, Kazuhiro Furukawa, Zasha Weinberg, Joy X Wang, Ronald R Breaker
Riboswitches in eubacteria sense the second messenger c-di-AMP.
Nat Chem Biol: 2013, 9(12);834-9
[PubMed:24141192]
[WorldCat.org]
[DOI]
(I p)
Ricardo S Vieira-Pires, Andras Szollosi, João H Morais-Cabral
The structure of the KtrAB potassium transporter.
Nature: 2013, 496(7445);323-8
[PubMed:23598340]
[WorldCat.org]
[DOI]
(I p)
Peter Y Watson, Martha J Fedor
The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis.
Nat Chem Biol: 2012, 8(12);963-5
[PubMed:23086297]
[WorldCat.org]
[DOI]
(I p)
Kirsten F Block, Ming C Hammond, Ronald R Breaker
Evidence for widespread gene control function by the ydaO riboswitch candidate.
J Bacteriol: 2010, 192(15);3983-9
[PubMed:20511502]
[WorldCat.org]
[DOI]
(I p)
Ronald A Albright, José-Luís Vazquez Ibar, Chae Un Kim, Sol M Gruner, João Henrique Morais-Cabral
The RCK domain of the KtrAB K+ transporter: multiple conformations of an octameric ring.
Cell: 2006, 126(6);1147-59
[PubMed:16990138]
[WorldCat.org]
[DOI]
(P p)
Rebecca F Kinsinger, Daniel B Kearns, Marina Hale, Ray Fall
Genetic requirements for potassium ion-dependent colony spreading in Bacillus subtilis.
J Bacteriol: 2005, 187(24);8462-9
[PubMed:16321950]
[WorldCat.org]
[DOI]
(P p)
Jeffrey E Barrick, Keith A Corbino, Wade C Winkler, Ali Nahvi, Maumita Mandal, Jennifer Collins, Mark Lee, Adam Roth, Narasimhan Sudarsan, Inbal Jona, J Kenneth Wickiser, Ronald R Breaker
New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control.
Proc Natl Acad Sci U S A: 2004, 101(17);6421-6
[PubMed:15096624]
[WorldCat.org]
[DOI]
(P p)
Gudrun Holtmann, Evert P Bakker, Nobuyuki Uozumi, Erhard Bremer
KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity.
J Bacteriol: 2003, 185(4);1289-98
[PubMed:12562800]
[WorldCat.org]
[DOI]
(P p)