Difference between revisions of "TnrA"
(→Database entries) |
(→Labs working on this gene/protein) |
||
| Line 147: | Line 147: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| − | + | * [[Susan Fisher]], Boston, USA [http://www.bumc.bu.edu/microbiology/research/susan-h-fisher-phd/ homepage] | |
| − | [[Susan Fisher]], Boston, USA [http://www.bumc.bu.edu/microbiology/research/susan-h-fisher-phd/ homepage] | + | * [[Maria Schumacher]], Durham, NC, USA [http://www.dukeschumacherlab.com/index.html homepage] |
=Your additional remarks= | =Your additional remarks= | ||
Revision as of 12:37, 5 March 2015
- Description: transcriptional pleiotropic regulator (MerR family) involved in global nitrogen regulation
| Gene name | tnrA |
| Synonyms | scgR |
| Essential | no |
| Product | transcription activator/ repressor (MerR family) |
| Function | regulation of nitrogen assimilation |
| Gene expression levels in SubtiExpress: tnrA | |
| Interactions involving this protein in SubtInteract: TnrA | |
| Metabolic function and regulation of this protein in SubtiPathways: tnrA | |
| MW, pI | 12 kDa, 10.235 |
| Gene length, protein length | 330 bp, 110 aa |
| Immediate neighbours | mgtE, ykzB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 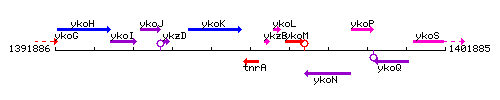
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, glutamate metabolism, transcription factors and their control, regulators of core metabolism
This gene is a member of the following regulons
The TnrA regulon
The gene
Basic information
- Locus tag: BSU13310
Phenotypes of a mutant
Database entries
- BsubCyc: BSU13310
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MerR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- K(D) value for the binding site in the tnrA promoter region: 55 nM PubMed
- Domains:
- Modification:
- Cofactor(s):
- Localization: membrane-associated via NrgA-NrgB under conditions of poor nitrogen supply PubMed
Database entries
- BsubCyc: BSU13310
- UniProt: Q45666
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: tnrA (according to DBTBS)
- Regulation:
- Additional information:
Biological materials
- Mutant: GP252 (in frame deletion), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Karl Forchhammer lab
Labs working on this gene/protein
- Susan Fisher, Boston, USA homepage
- Maria Schumacher, Durham, NC, USA homepage
Your additional remarks
References
Reviews
Katrin Gunka, Fabian M Commichau
Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation.
Mol Microbiol: 2012, 85(2);213-24
[PubMed:22625175]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Jörg Stülke
Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression.
Mol Microbiol: 2008, 67(4);692-702
[PubMed:18086213]
[WorldCat.org]
[DOI]
(P p)
S H Fisher
Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence!
Mol Microbiol: 1999, 32(2);223-32
[PubMed:10231480]
[WorldCat.org]
[DOI]
(P p)
The TnrA regulon
Control of TnrA activity by the trigger enzyme GlnA
Other original publications