Difference between revisions of "YusV"
| Line 39: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 106: | Line 102: | ||
* '''Structure:''' | * '''Structure:''' | ||
| + | ** [http://www.pdb.org/pdb/explore/explore.do?structureId=4R9U 4R9U], the E. coli BtuC-BtuD complex, BtuD shares 32% identity, 57% similarity with YusV, {{PubMed|25402482}} | ||
* '''UniProt:''' [http://www.uniprot.org/uniprot/O32188 O32188] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O32188 O32188] | ||
| Line 153: | Line 150: | ||
=References= | =References= | ||
| − | <pubmed>19746494,10092453,16672620,12354229, </pubmed> | + | <pubmed>19746494,10092453,16672620,12354229, 25402482 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:41, 26 November 2014
- Description: ABC transporter for the siderophores Fe-enterobactin and Fe-bacillibactin, as well as for the siderophores schizokinen and arthrobactin (ATPase)
| Gene name | yusV |
| Synonyms | |
| Essential | no |
| Product | ABC transporter for the siderophores Fe-enterobactin, Fe-bacillibactin, schizokinen and arthrobactin (ATPase) |
| Function | acquisition of iron |
| Gene expression levels in SubtiExpress: yusV | |
| Interactions involving this protein in SubtInteract: YusV | |
| Metabolic function and regulation of this protein in SubtiPathways: YusV | |
| MW, pI | 30 kDa, 5.322 |
| Gene length, protein length | 825 bp, 275 aa |
| Immediate neighbours | yusU, yusW |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 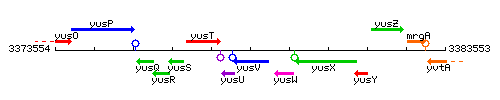
This image was kindly provided by SubtiList
| |
| Expression at a glance PubMed 500px | |
Contents
Categories containing this gene/protein
ABC transporters, acquisition of iron, iron metabolism, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32940
Phenotypes of a mutant
Database entries
- BsubCyc: BSU32940
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase for the FeuA-FeuB-FeuC siderophore ABC transporter and for the YfhA-YfiY-YfiZ siderophore ABC transporter
- Protein family: ABC transporter family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU32940
- Structure:
- UniProt: O32188
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References