Difference between revisions of "MurAA"
(→Biological materials) |
|||
| Line 58: | Line 58: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU36760&redirect=T BSU36760] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/murAA.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/murAA.html] | ||
| Line 95: | Line 96: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU36760&redirect=T BSU36760] | ||
* '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=3SG1 3SG1] (from ''B. anthracis'', 79% identity, 94% similarity) | * '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=3SG1 3SG1] (from ''B. anthracis'', 79% identity, 94% similarity) | ||
Revision as of 15:02, 2 April 2014
- Description: UDP-N-acetylglucosamine 1-carboxyvinyltransferase
| Gene name | murAA |
| Synonyms | murA |
| Essential | yes PubMed |
| Product | UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
| Function | peptidoglycan precursor biosynthesis |
| Gene expression levels in SubtiExpress: murAA | |
| Metabolic function and regulation of this protein in SubtiPathways: murAA | |
| MW, pI | 46 kDa, 5.45 |
| Gene length, protein length | 1308 bp, 436 aa |
| Immediate neighbours | spoIID, ywmB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 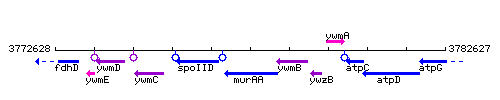
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed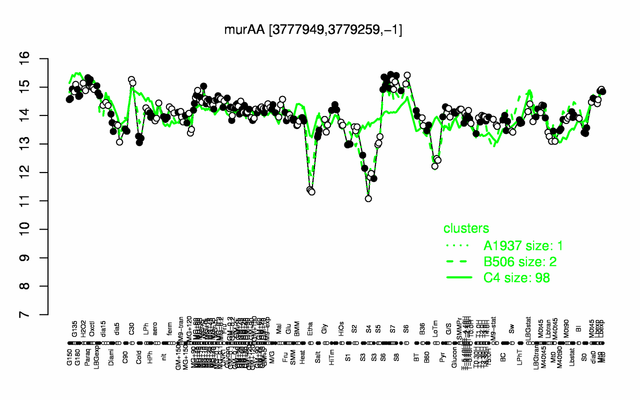
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU36760
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU36760
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Phosphoenolpyruvate + UDP-N-acetyl-D-glucosamine = phosphate + UDP-N-acetyl-3-O-(1-carboxyvinyl)-D-glucosamine (according to Swiss-Prot)
- Protein family: MurA subfamily (according to Swiss-Prot)
- Paralogous protein(s): MurAB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU36760
- Structure: 3SG1 (from B. anthracis, 79% identity, 94% similarity)
- UniProt: P70965
- KEGG entry: [3]
- E.C. number: 2.5.1.7
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- pRSETA-murAA, Expression of His(6)-MurAA, avalable in Ulf Gerth's lab PubMed
- pGP2597: (IPTG inducible expression, purification in E. coli with N-terminal Strep-tag, in pGP172), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Sean G Jackson, Fuzhong Zhang, Paul Chindemi, Murray S Junop, Paul J Berti
Evidence of kinetic control of ligand binding and staged product release in MurA (enolpyruvyl UDP-GlcNAc synthase)-catalyzed reactions .
Biochemistry: 2009, 48(49);11715-23
[PubMed:19899805]
[WorldCat.org]
[DOI]
(I p)
Jean van Heijenoort
Lipid intermediates in the biosynthesis of bacterial peptidoglycan.
Microbiol Mol Biol Rev: 2007, 71(4);620-35
[PubMed:18063720]
[WorldCat.org]
[DOI]
(P p)
Holger Kock, Ulf Gerth, Michael Hecker
MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis.
Mol Microbiol: 2004, 51(4);1087-102
[PubMed:14763982]
[WorldCat.org]
[DOI]
(P p)
T Skarzynski, D H Kim, W J Lees, C T Walsh, K Duncan
Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA.
Biochemistry: 1998, 37(8);2572-7
[PubMed:9485407]
[WorldCat.org]
[DOI]
(P p)
T Skarzynski, A Mistry, A Wonacott, S E Hutchinson, V A Kelly, K Duncan
Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Structure: 1996, 4(12);1465-74
[PubMed:8994972]
[WorldCat.org]
[DOI]
(P p)