Difference between revisions of "YwrJ"
| Line 56: | Line 56: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU36040&redirect=T BSU36040] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ywrJ.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ywrJ.html] | ||
| Line 93: | Line 94: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU36040&redirect=T BSU36040] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 14:59, 2 April 2014
- Description: sporulation protein
| Gene name | ywrJ |
| Synonyms | |
| Essential | no |
| Product | sporulation protein |
| Function | unknown |
| Gene expression levels in SubtiExpress: ywrJ | |
| MW, pI | 25 kDa, 5.957 |
| Gene length, protein length | 675 bp, 225 aa |
| Immediate neighbours | ywrK, cotB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 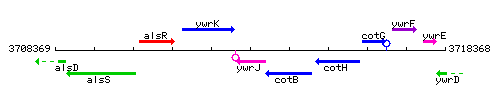
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed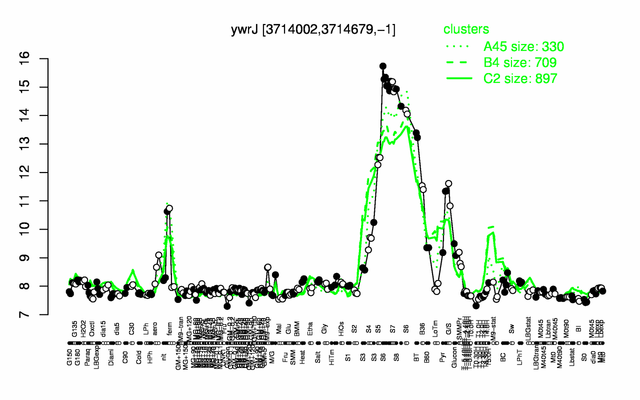
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU36040
Phenotypes of a mutant
Database entries
- BsubCyc: BSU36040
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on Ser-208 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- BsubCyc: BSU36040
- Structure:
- UniProt: O05223
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References