Difference between revisions of "FtsA"
| Line 60: | Line 60: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15280&redirect=T BSU15280] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ftsAZ.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ftsAZ.html] | ||
| Line 98: | Line 99: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15280&redirect=T BSU15280] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 13:39, 2 April 2014
- Description: cell division protein, membrane anchor for FtsZ
| Gene name | ftsA |
| Synonyms | spoIIN |
| Essential | no |
| Product | cell division protein |
| Function | formation of Z-ring |
| Gene expression levels in SubtiExpress: ftsA | |
| Interactions involving this protein in SubtInteract: FtsA | |
| MW, pI | 47 kDa, 5.094 |
| Gene length, protein length | 1320 bp, 440 aa |
| Immediate neighbours | sbp, ftsZ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 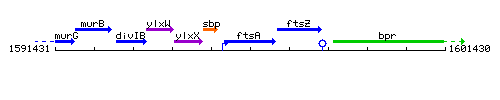
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed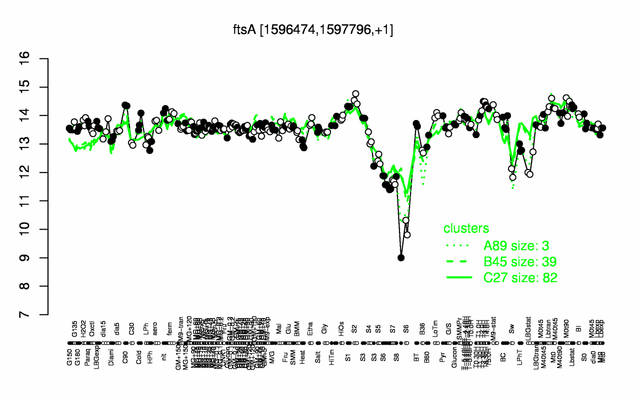
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15280
Phenotypes of a mutant
Database entries
- BsubCyc: BSU15280
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ftsA/mreB family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU15280
- Structure:
- UniProt: P28264
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Martin Loose, Timothy J Mitchison
The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns.
Nat Cell Biol: 2014, 16(1);38-46
[PubMed:24316672]
[WorldCat.org]
[DOI]
(I p)
Hiraku Takada, Sanae Fukushima-Tanaka, Masato Morita, Yasuhiro Kasahara, Satoru Watanabe, Taku Chibazakura, Hiroshi Hara, Kouji Matsumoto, Hirofumi Yoshikawa
An essential enzyme for phospholipid synthesis associates with the Bacillus subtilis divisome.
Mol Microbiol: 2014, 91(2);242-55
[PubMed:24224907]
[WorldCat.org]
[DOI]
(I p)
Ramona Duman, Shu Ishikawa, Ilkay Celik, Henrik Strahl, Naotake Ogasawara, Paulina Troc, Jan Löwe, Leendert W Hamoen
Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring.
Proc Natl Acad Sci U S A: 2013, 110(48);E4601-10
[PubMed:24218584]
[WorldCat.org]
[DOI]
(I p)
Erik Nico Trip, Jan-Willem Veening, Eric J Stewart, Jeff Errington, Dirk-Jan Scheffers
Balanced transcription of cell division genes in Bacillus subtilis as revealed by single cell analysis.
Environ Microbiol: 2013, 15(12);3196-209
[PubMed:23701187]
[WorldCat.org]
[DOI]
(I p)
Parminder Singh, Ravindra D Makde, Saikat Ghosh, Jayant Asthana, Vinay Kumar, Dulal Panda
Assembly of Bacillus subtilis FtsA: effects of pH, ionic strength and nucleotides on FtsA assembly.
Int J Biol Macromol: 2013, 52;170-6
[PubMed:23036588]
[WorldCat.org]
[DOI]
(I p)
Henrik Strahl, Leendert W Hamoen
Membrane potential is important for bacterial cell division.
Proc Natl Acad Sci U S A: 2010, 107(27);12281-6
[PubMed:20566861]
[WorldCat.org]
[DOI]
(I p)
José Roberto Tavares, Robson F de Souza, Guilherme Louzada Silva Meira, Frederico J Gueiros-Filho
Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome.
J Bacteriol: 2008, 190(21);7096-107
[PubMed:18776011]
[WorldCat.org]
[DOI]
(I p)
Shu Ishikawa, Yoshikazu Kawai, Konosuke Hiramatsu, Masayoshi Kuwano, Naotake Ogasawara
A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis.
Mol Microbiol: 2006, 60(6);1364-80
[PubMed:16796675]
[WorldCat.org]
[DOI]
(P p)
S O Jensen, L S Thompson, E J Harry
Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-Ring assembly.
J Bacteriol: 2005, 187(18);6536-44
[PubMed:16159787]
[WorldCat.org]
[DOI]
(P p)
Jennifer T Kemp, Adam Driks, Richard Losick
FtsA mutants of Bacillus subtilis impaired in sporulation.
J Bacteriol: 2002, 184(14);3856-63
[PubMed:12081956]
[WorldCat.org]
[DOI]
(P p)
A Feucht, I Lucet, M D Yudkin, J Errington
Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis.
Mol Microbiol: 2001, 40(1);115-25
[PubMed:11298280]
[WorldCat.org]
[DOI]
(P p)
Keisuke Fukuchi, Yasuhiro Kasahara, Kei Asai, Kazuo Kobayashi, Shigeki Moriya, Naotake Ogasawara
The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis.
Microbiology (Reading): 2000, 146 ( Pt 7);1573-1583
[PubMed:10878122]
[WorldCat.org]
[DOI]
(P p)
X Wang, J Huang, A Mukherjee, C Cao, J Lutkenhaus
Analysis of the interaction of FtsZ with itself, GTP, and FtsA.
J Bacteriol: 1997, 179(17);5551-9
[PubMed:9287012]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promote.
J Bacteriol: 1995, 177(23);6999-7002
[PubMed:7592498]
[WorldCat.org]
[DOI]
(P p)
G Gonzy-Tréboul, C Karmazyn-Campelli, P Stragier
Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon.
J Mol Biol: 1992, 224(4);967-79
[PubMed:1569582]
[WorldCat.org]
[DOI]
(P p)