Difference between revisions of "YhdN"
(→The gene) |
|||
| Line 56: | Line 56: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU09530&redirect=T BSU09530] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yhdNO.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yhdNO.html] | ||
| Line 91: | Line 92: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU09530&redirect=T BSU09530] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1PZ1 1PZ1] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1PZ1 1PZ1] | ||
Revision as of 13:18, 2 April 2014
- Description: general stress protein, broad specificity aldo-keto reductase that converts MG to acetol
| Gene name | yhdN |
| Synonyms | |
| Essential | no |
| Product | aldo/keto reductase, specific for NADP |
| Function | detoxification of methylglyoxal |
| Gene expression levels in SubtiExpress: yhdN | |
| Metabolic function and regulation of this protein in SubtiPathways: yhdN | |
| MW, pI | 37 kDa, 4.77 |
| Gene length, protein length | 993 bp, 331 aa |
| Immediate neighbours | sigM, plsC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 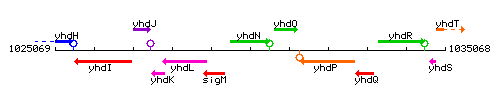
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed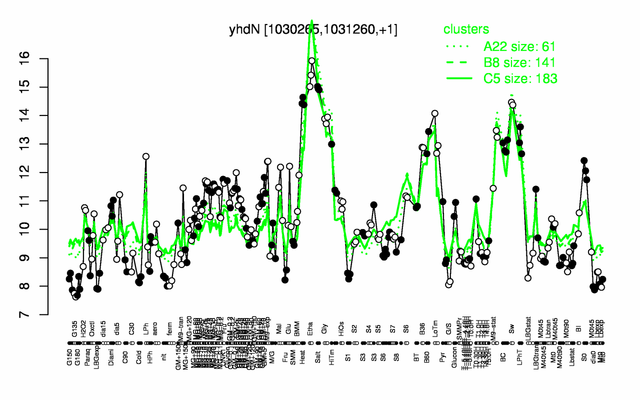
| |
Contents
Categories containing this gene/protein
biosynthesis of lipids, general stress proteins (controlled by SigB), resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU09530
Phenotypes of a mutant
- increased sensitivity to methylglyoxal PubMed
Database entries
- BsubCyc: BSU09530
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- methylglyoxal --> acetol PubMed
- Protein family: aldo/keto reductase 2 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU09530
- Structure: 1PZ1
- UniProt: P80874
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Pete Chandrangsu, Renata Dusi, Chris J Hamilton, John D Helmann
Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways.
Mol Microbiol: 2014, 91(4);706-15
[PubMed:24330391]
[WorldCat.org]
[DOI]
(I p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Andreas H Ehrensberger, David K Wilson
Structural and catalytic diversity in the two family 11 aldo-keto reductases.
J Mol Biol: 2004, 337(3);661-73
[PubMed:15019785]
[WorldCat.org]
[DOI]
(P p)
Gustavo E Schujman, Luciana Paoletti, Alan D Grossman, Diego de Mendoza
FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis.
Dev Cell: 2003, 4(5);663-72
[PubMed:12737802]
[WorldCat.org]
[DOI]
(P p)
Anja Petersohn, Haike Antelmann, Ulf Gerth, Michael Hecker
Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 4);869-880
[PubMed:10220166]
[WorldCat.org]
[DOI]
(P p)