Difference between revisions of "PurE"
| Line 58: | Line 58: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU06420&redirect=T BSU06420] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/purEKBCSQLFMNHD.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/purEKBCSQLFMNHD.html] | ||
| Line 95: | Line 96: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU06420&redirect=T BSU06420] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1O4V 1O4V] (from ''Thermotoga maritima'', 53% identity, 64% similarity) {{PubMed|15048837}} | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1O4V 1O4V] (from ''Thermotoga maritima'', 53% identity, 64% similarity) {{PubMed|15048837}} | ||
Revision as of 13:07, 2 April 2014
- Description: phosphoribosylaminoimidazole carboxylase (ATP-dependent)
| Gene name | purE |
| Synonyms | |
| Essential | no |
| Product | phosphoribosylaminoimidazole carboxylase (ATP-dependent) |
| Function | purine biosynthesis |
| Gene expression levels in SubtiExpress: purE | |
| Metabolic function and regulation of this protein in SubtiPathways: purE | |
| MW, pI | 17 kDa, 5.199 |
| Gene length, protein length | 486 bp, 162 aa |
| Immediate neighbours | yebG, purK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 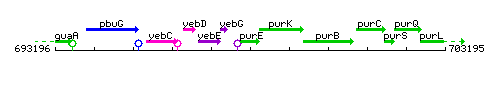
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06420
Phenotypes of a mutant
Database entries
- BsubCyc: BSU06420
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate = 5-amino-1-(5-phospho-D-ribosyl)imidazole + CO2 (according to Swiss-Prot)
- Protein family: AIR carboxylase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU06420
- UniProt: P12044
- KEGG entry: [3]
- E.C. number: 4.1.1.21
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
Maumita Mandal, Benjamin Boese, Jeffrey E Barrick, Wade C Winkler, Ronald R Breaker
Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria.
Cell: 2003, 113(5);577-86
[PubMed:12787499]
[WorldCat.org]
[DOI]
(P p)
M Weng, P L Nagy, H Zalkin
Identification of the Bacillus subtilis pur operon repressor.
Proc Natl Acad Sci U S A: 1995, 92(16);7455-9
[PubMed:7638212]
[WorldCat.org]
[DOI]
(P p)
D J Ebbole, H Zalkin
Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis.
J Biol Chem: 1987, 262(17);8274-87
[PubMed:3036807]
[WorldCat.org]
(P p)