Difference between revisions of "GsiB"
| Line 53: | Line 53: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU04400&redirect=T BSU04400] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gsiB.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gsiB.html] | ||
| Line 90: | Line 91: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU04400&redirect=T BSU04400] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 13:01, 2 April 2014
- Description: general stress protein, prevents enzyme inactivation upon freeze-thaw treatments
| Gene name | gsiB |
| Synonyms | |
| Essential | no |
| Product | general stress protein |
| Function | response to water deficits |
| Gene expression levels in SubtiExpress: gsiB | |
| MW, pI | 13 kDa, 5.145 |
| Gene length, protein length | 369 bp, 123 aa |
| Immediate neighbours | ydbA, ydbB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 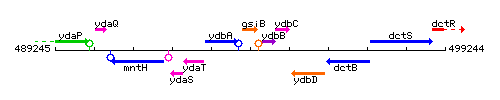
This image was kindly provided by SubtiList
| |
[None Expression at a glance] PubMed
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04400
Phenotypes of a mutant
Database entries
- BsubCyc: BSU04400
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU04400
- Structure:
- UniProt: P26907
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: gsiB PubMed
- Regulatory mechanism:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
F Campos, C Cuevas-Velazquez, M A Fares, J L Reyes, A A Covarrubias
Group 1 LEA proteins, an ancestral plant protein group, are also present in other eukaryotes, and in the archeae and bacteria domains.
Mol Genet Genomics: 2013, 288(10);503-17
[PubMed:23861025]
[WorldCat.org]
[DOI]
(I p)
Ioanna-Areti Asteri, Effrossyni Boutou, Rania Anastasiou, Bruno Pot, Constantinos E Vorgias, Effie Tsakalidou, Konstantinos Papadimitriou
In silico evidence for the horizontal transfer of gsiB, a σ(B)-regulated gene in gram-positive bacteria, to lactic acid bacteria.
Appl Environ Microbiol: 2011, 77(10);3526-31
[PubMed:21421783]
[WorldCat.org]
[DOI]
(I p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
B Jürgen, T Schweder, M Hecker
The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site.
Mol Gen Genet: 1998, 258(5);538-45
[PubMed:9669336]
[WorldCat.org]
[DOI]
(P p)
B Maul, U Völker, S Riethdorf, S Engelmann, M Hecker
sigma B-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis.
Mol Gen Genet: 1995, 248(1);114-20
[PubMed:7651322]
[WorldCat.org]
[DOI]
(P p)
J P Mueller, G Bukusoglu, A L Sonenshein
Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system.
J Bacteriol: 1992, 174(13);4361-73
[PubMed:1378051]
[WorldCat.org]
[DOI]
(P p)