Difference between revisions of "PpaC"
| Line 105: | Line 105: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''ppaC'' | + | * '''Operon:''' ''ppaC'' {{PubMed|22383849}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ppaC_4168204_4169133_1 ppaC] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ppaC_4168204_4169133_1 ppaC] {{PubMed|22383849}} | ||
| Line 138: | Line 138: | ||
=References= | =References= | ||
| − | <pubmed>22938038,21749987,15533045,17611193,9845334 9782505 11342544, 9633597 21749987 15378759</pubmed> | + | <pubmed>22938038,21749987,15533045,17611193,9845334 9782505 11342544, 9633597 21749987 15378759 22383849</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:50, 1 April 2014
- Description: inorganic pyrophosphatase
| Gene name | ppaC |
| Synonyms | yybQ |
| Essential | yes PubMed |
| Product | inorganic pyrophosphatase |
| Function | recovery of phosphate ions from pyrophosphate |
| Gene expression levels in SubtiExpress: ppaC | |
| MW, pI | 33 kDa, 4.513 |
| Gene length, protein length | 927 bp, 309 aa |
| Immediate neighbours | hypR, yybP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 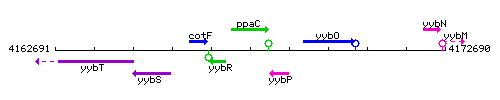
This image was kindly provided by SubtiList
| |
| Expression at a glance PubMed 500px | |
Contents
Categories containing this gene/protein
phosphate metabolism, essential genes, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40550
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Diphosphate + H2O = 2 phosphate (according to Swiss-Prot)
- Protein family: PPase class C family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: P37487
- KEGG entry: [3]
- E.C. number: 3.6.1.1
Additional information
Expression and regulation
- Operon: ppaC PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Bui Khanh Chi, Alexandra A Roberts, Tran Thi Thanh Huyen, Katrin Bäsell, Dörte Becher, Dirk Albrecht, Chris J Hamilton, Haike Antelmann
S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria.
Antioxid Redox Signal: 2013, 18(11);1273-95
[PubMed:22938038]
[WorldCat.org]
[DOI]
(I p)
Pierre Nicolas, Ulrike Mäder, Etienne Dervyn, Tatiana Rochat, Aurélie Leduc, Nathalie Pigeonneau, Elena Bidnenko, Elodie Marchadier, Mark Hoebeke, Stéphane Aymerich, Dörte Becher, Paola Bisicchia, Eric Botella, Olivier Delumeau, Geoff Doherty, Emma L Denham, Mark J Fogg, Vincent Fromion, Anne Goelzer, Annette Hansen, Elisabeth Härtig, Colin R Harwood, Georg Homuth, Hanne Jarmer, Matthieu Jules, Edda Klipp, Ludovic Le Chat, François Lecointe, Peter Lewis, Wolfram Liebermeister, Anika March, Ruben A T Mars, Priyanka Nannapaneni, David Noone, Susanne Pohl, Bernd Rinn, Frank Rügheimer, Praveen K Sappa, Franck Samson, Marc Schaffer, Benno Schwikowski, Leif Steil, Jörg Stülke, Thomas Wiegert, Kevin M Devine, Anthony J Wilkinson, Jan Maarten van Dijl, Michael Hecker, Uwe Völker, Philippe Bessières, Philippe Noirot
Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis.
Science: 2012, 335(6072);1103-6
[PubMed:22383849]
[WorldCat.org]
[DOI]
(I p)
Bui Khanh Chi, Katrin Gronau, Ulrike Mäder, Bernd Hessling, Dörte Becher, Haike Antelmann
S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics.
Mol Cell Proteomics: 2011, 10(11);M111.009506
[PubMed:21749987]
[WorldCat.org]
[DOI]
(I p)
Falko Hochgräfe, Jörg Mostertz, Dierk-Christoph Pöther, Dörte Becher, John D Helmann, Michael Hecker
S-cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress.
J Biol Chem: 2007, 282(36);25981-5
[PubMed:17611193]
[WorldCat.org]
[DOI]
(P p)
Igor P Fabrichniy, Lari Lehtiö, Anu Salminen, Anton B Zyryanov, Alexander A Baykov, Reijo Lahti, Adrian Goldman
Structural studies of metal ions in family II pyrophosphatases: the requirement for a Janus ion.
Biochemistry: 2004, 43(45);14403-11
[PubMed:15533045]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
A N Parfenyev, A Salminen, P Halonen, A Hachimori, A A Baykov, R Lahti
Quaternary structure and metal ion requirement of family II pyrophosphatases from Bacillus subtilis, Streptococcus gordonii, and Streptococcus mutans.
J Biol Chem: 2001, 276(27);24511-8
[PubMed:11342544]
[WorldCat.org]
[DOI]
(P p)
T Shintani, T Uchiumi, T Yonezawa, A Salminen, A A Baykov, R Lahti, A Hachimori
Cloning and expression of a unique inorganic pyrophosphatase from Bacillus subtilis: evidence for a new family of enzymes.
FEBS Lett: 1998, 439(3);263-6
[PubMed:9845334]
[WorldCat.org]
[DOI]
(P p)
Tom W Young, Nicholas J Kuhn, Albert Wadeson, Simon Ward, Dan Burges, G Dunstan Cooke
Bacillus subtilis ORF yybQ encodes a manganese-dependent inorganic pyrophosphatase with distinctive properties: the first of a new class of soluble pyrophosphatase?
Microbiology (Reading): 1998, 144 ( Pt 9);2563-2571
[PubMed:9782505]
[WorldCat.org]
[DOI]
(P p)
N J Kuhn, S Ward
Purification, properties, and multiple forms of a manganese-activated inorganic pyrophosphatase from Bacillus subtilis.
Arch Biochem Biophys: 1998, 354(1);47-56
[PubMed:9633597]
[WorldCat.org]
[DOI]
(P p)