Difference between revisions of "ScpB"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' part of the [[condensin]] complex, chromosomal origin condensation and segregation <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || DNA segregation and condensation protein | |style="background:#ABCDEF;" align="center"| '''Product''' || DNA segregation and condensation protein | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || segregation of replication origins |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23210 scpB] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23210 scpB] | ||
| Line 52: | Line 52: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | * | + | * ''[[scpB]]'' mutants are not viable on complex medim that allow rapid growth, but they are viable under conditions of slow growth {{PubMed|24440399,24440393}} |
=== Database entries === | === Database entries === | ||
| Line 76: | Line 76: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** part of the [[condensin]] complex {{PubMed|24440399,24440393}} | ||
** forms dimers {{PubMed|22385855}} | ** forms dimers {{PubMed|22385855}} | ||
** [[ScpB]]-[[Smc]] {{PubMed|22385855,12100548}}, this interaction takes place only in the presence of [[ScpA]] {{PubMed|22385855}} | ** [[ScpB]]-[[Smc]] {{PubMed|22385855,12100548}}, this interaction takes place only in the presence of [[ScpA]] {{PubMed|22385855}} | ||
| Line 91: | Line 92: | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
** nucleoid (Multiple) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | ** nucleoid (Multiple) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | ||
| − | ** the ([[Smc]])2-[[ScpA]]-[[ScpB]] complex forms two bipolar assemblies on the chromosome, one in each cell half {{PubMed|23475963}} | + | ** the [[condensin]] ([[Smc]])2-[[ScpA]]-[[ScpB]] complex forms two bipolar assemblies on the chromosome, one in each cell half, localization depends on [[Spo0J]] {{PubMed|24440393,23475963}} |
=== Database entries === | === Database entries === | ||
| Line 143: | Line 144: | ||
<pubmed> 22933559 22934648 </pubmed> | <pubmed> 22933559 22934648 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>12100548,12065423,12421306,22385855, 16479537, 19450516 12897137 7934830 11948165 23353789 24440393 | + | <pubmed>12100548,12065423,12421306,22385855, 16479537, 19450516 12897137 7934830 11948165 23353789 24440399,24440393 23475963</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:28, 16 February 2014
- Description: part of the condensin complex, chromosomal origin condensation and segregation
| Gene name | scpB |
| Synonyms | ypuH |
| Essential | no |
| Product | DNA segregation and condensation protein |
| Function | segregation of replication origins |
| Gene expression levels in SubtiExpress: scpB | |
| Interactions involving this protein in SubtInteract: ScpB | |
| MW, pI | 21 kDa, 4.25 |
| Gene length, protein length | 591 bp, 197 aa |
| Immediate neighbours | ypuI, scpA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 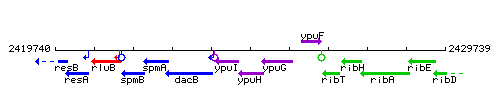
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23210
Phenotypes of a mutant
- scpB mutants are not viable on complex medim that allow rapid growth, but they are viable under conditions of slow growth PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: scpB family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- UniProt: P35155
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Stephan Gruber, Jan-Willem Veening, Juri Bach, Martin Blettinger, Marc Bramkamp, Jeff Errington
Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis.
Curr Biol: 2014, 24(3);293-8
[PubMed:24440399]
[WorldCat.org]
[DOI]
(I p)
Xindan Wang, Olive W Tang, Eammon P Riley, David Z Rudner
The SMC condensin complex is required for origin segregation in Bacillus subtilis.
Curr Biol: 2014, 24(3);287-92
[PubMed:24440393]
[WorldCat.org]
[DOI]
(I p)
Luise A K Kleine Borgmann, Hanna Hummel, Maximilian H Ulbrich, Peter L Graumann
SMC condensation centers in Bacillus subtilis are dynamic structures.
J Bacteriol: 2013, 195(10);2136-45
[PubMed:23475963]
[WorldCat.org]
[DOI]
(I p)
Frank Bürmann, Ho-Chul Shin, Jérôme Basquin, Young-Min Soh, Victor Giménez-Oya, Yeon-Gil Kim, Byung-Ha Oh, Stephan Gruber
An asymmetric SMC-kleisin bridge in prokaryotic condensin.
Nat Struct Mol Biol: 2013, 20(3);371-9
[PubMed:23353789]
[WorldCat.org]
[DOI]
(I p)
M E Fuentes-Perez, E J Gwynn, M S Dillingham, F Moreno-Herrero
Using DNA as a fiducial marker to study SMC complex interactions with the atomic force microscope.
Biophys J: 2012, 102(4);839-48
[PubMed:22385855]
[WorldCat.org]
[DOI]
(I p)
Stephan Gruber, Jeff Errington
Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis.
Cell: 2009, 137(4);685-96
[PubMed:19450516]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
A Volkov, J Mascarenhas, C Andrei-Selmer, H D Ulrich, P L Graumann
A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure.
Mol Cell Biol: 2003, 23(16);5638-50
[PubMed:12897137]
[WorldCat.org]
[DOI]
(P p)
Janet C Lindow, Masayoshi Kuwano, Shigeki Moriya, Alan D Grossman
Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein.
Mol Microbiol: 2002, 46(4);997-1009
[PubMed:12421306]
[WorldCat.org]
[DOI]
(P p)
Jörg Soppa, Kazuo Kobayashi, Marie-Françoise Noirot-Gros, Dieter Oesterhelt, S Dusko Ehrlich, Etienne Dervyn, Naotake Ogasawara, Shigeki Moriya
Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB.
Mol Microbiol: 2002, 45(1);59-71
[PubMed:12100548]
[WorldCat.org]
[DOI]
(P p)
Judita Mascarenhas, Jörg Soppa, Alexander V Strunnikov, Peter L Graumann
Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein.
EMBO J: 2002, 21(12);3108-18
[PubMed:12065423]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
V Azevedo, A Sorokin, S D Ehrlich, P Serror
The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci.
Mol Microbiol: 1993, 10(2);397-405
[PubMed:7934830]
[WorldCat.org]
[DOI]
(P p)