Difference between revisions of "DegU"
(→Phenotypes of a mutant) |
|||
| Line 59: | Line 59: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | * defect in [[biofilm formation]] {{PubMed|20815827}}, this can be suppressed by DegU-independent expression of [[ | + | * defect in [[biofilm formation]] {{PubMed|20815827}}, this can be suppressed by [[DegU]]-independent expression of [[BslA]] {{PubMed|21742882}} |
* the mutation suppresses the mucoid phenotype of ''[[motA]]'' or ''[[motB]]'' mutants due to reduced expression of the ''[[capB]]-[[capC]]-[[capA]]-[[capE]]'' operon {{PubMed|24296669}} | * the mutation suppresses the mucoid phenotype of ''[[motA]]'' or ''[[motB]]'' mutants due to reduced expression of the ''[[capB]]-[[capC]]-[[capA]]-[[capE]]'' operon {{PubMed|24296669}} | ||
| Line 113: | Line 113: | ||
=== Additional information=== | === Additional information=== | ||
* [[DegU]]-P is degraded by [[ClpC]]-[[ClpP]] {{PubMed|20070525}} | * [[DegU]]-P is degraded by [[ClpC]]-[[ClpP]] {{PubMed|20070525}} | ||
| + | * accumulation of [[DegU]]-P results in decreased transcription of the ''[[epsA]]-[[epsO]]'' and ''[[tapA]]-[[sipW]]-[[tasA]]'' operons due to increased levels of [[Spo0A]]-P {{PubMed|24123822}} | ||
| + | |||
=Expression and regulation= | =Expression and regulation= | ||
Revision as of 18:50, 15 February 2014
- Description: two-component response regulator, regulation of degradative enzyme and genetic competence, involved in activation of capsule biosynthetic operon expression (together with SwrAA), non-phosphorylated DegU is required for swarming motility PubMed
| Gene name | degU |
| Synonyms | sacU, iep |
| Essential | no |
| Product | two-component response regulator |
| Function | regulation of degradative enzyme and genetic competence |
| Gene expression levels in SubtiExpress: degU | |
| Interactions involving this protein in SubtInteract: DegU | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 25 kDa, 5.602 |
| Gene length, protein length | 687 bp, 229 aa |
| Immediate neighbours | yviA, degS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 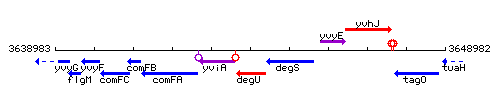
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biofilm formation, transcription factors and their control, phosphoproteins
This gene is a member of the following regulons
CcpA regulon, DegU regulon, TnrA regulon
The DegU regulon
The gene
Basic information
- Locus tag: BSU35490
Phenotypes of a mutant
- defect in biofilm formation PubMed, this can be suppressed by DegU-independent expression of BslA PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants due to reduced expression of the capB-capC-capA-capE operon PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activator of degradative enzyme genes (sacB and degQ) and of comK expression
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P13800
- KEGG entry: [3]
- E.C. number:
Additional information
- DegU-P is degraded by ClpC-ClpP PubMed
- accumulation of DegU-P results in decreased transcription of the epsA-epsO and tapA-sipW-tasA operons due to increased levels of Spo0A-P PubMed
Expression and regulation
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The DegU regulon
U Mäder, H Antelmann, T Buder, M K Dahl, M Hecker, G Homuth
Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics.
Mol Genet Genomics: 2002, 268(4);455-67
[PubMed:12471443]
[WorldCat.org]
[DOI]
(P p)
Original Publications