Difference between revisions of "RecU"
| Line 1: | Line 1: | ||
| − | * '''Description:''' DNA repair, homologous recombination and chromosome segregation; recognizes, distorts, and cleaves four-stranded recombination intermediates and modulates [[RecA]] activities <br/><br/> | + | * '''Description:''' DNA repair, homologous recombination and chromosome segregation; recognizes, distorts, and cleaves four-stranded recombination intermediates and modulates [[RecA]] activities, equivalent to ''E. coli'' RecU<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 79: | Line 79: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 112: | Line 112: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=recU_2340802_2341422_1 recU] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=recU_2340802_2341422_1 recU] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigM]] {{PubMed|18179421}} | + | * '''[[Sigma factor]]:''' [[SigM]] {{PubMed|18179421}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 123: | Line 123: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
| − | ** GP891 (''recU''::''cat''), available in [[Stülke]] lab | + | ** GP891 (''recU''::''cat''), available in [[Jörg Stülke]]'s lab |
** 1A895 (no resistance), {{PubMed|16020779}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A895&Search=1A895 BGSC] | ** 1A895 (no resistance), {{PubMed|16020779}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A895&Search=1A895 BGSC] | ||
| Line 142: | Line 142: | ||
=References= | =References= | ||
== Reviews == | == Reviews == | ||
| − | <pubmed> 22933559 </pubmed> | + | <pubmed> 22933559 9442895</pubmed> |
== Original publications == | == Original publications == | ||
<pubmed>9642195,16024744,15317759,11810266,19730681 , 19422832 7814321 18179421 16154091 21600217 18684995 16020779 </pubmed> | <pubmed>9642195,16024744,15317759,11810266,19730681 , 19422832 7814321 18179421 16154091 21600217 18684995 16020779 </pubmed> | ||
Revision as of 17:56, 14 December 2013
- Description: DNA repair, homologous recombination and chromosome segregation; recognizes, distorts, and cleaves four-stranded recombination intermediates and modulates RecA activities, equivalent to E. coli RecU
| Gene name | recU |
| Synonyms | recG, prfA, yppB, jopB |
| Essential | no |
| Product | Holliday junction resolvase |
| Function | DNA repair, homologous recombination and chromosome segregation |
| Gene expression levels in SubtiExpress: recU | |
| Interactions involving this protein in SubtInteract: RecU | |
| MW, pI | 23 kDa, 9.511 |
| Gene length, protein length | 618 bp, 206 aa |
| Immediate neighbours | yppC, ponA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 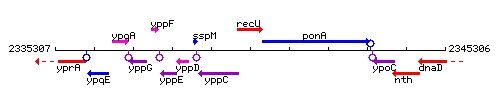
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure: 1ZP7
- UniProt: P39792
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP891 (recU::cat), available in Jörg Stülke's lab
- 1A895 (no resistance), PubMed, available at BGSC
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
RecU in other organisms
Ana R Pereira, Patricia Reed, Helena Veiga, Mariana G Pinho
The Holliday junction resolvase RecU is required for chromosome segregation and DNA damage repair in Staphylococcus aureus.
BMC Microbiol: 2013, 13;18
[PubMed:23356868]
[WorldCat.org]
[DOI]
(I e)