Difference between revisions of "MetE"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 81: | Line 77: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| Line 89: | Line 85: | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 116: | Line 112: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=metE_1383320_1385608_-1 metE] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=metE_1383320_1385608_-1 metE] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 146: | Line 142: | ||
=References= | =References= | ||
| − | + | <pubmed>21749987 22938038,21749987,19258532,16194229,10094622,17611193,18039762 ,17726680 12107147 24313874 </pubmed> | |
| − | <pubmed>22938038,21749987,19258532,16194229,10094622,17611193,18039762 ,17726680 12107147</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:52, 13 December 2013
- Description: methionine synthase
| Gene name | metE |
| Synonyms | metC |
| Essential | no |
| Product | methionine synthase |
| Function | biosynthesis of methionine |
| Gene expression levels in SubtiExpress: metE | |
| Metabolic function and regulation of this protein in SubtiPathways: Cys, Met & Sulfate assimilation | |
| MW, pI | 86 kDa, 4.839 |
| Gene length, protein length | 2286 bp, 762 aa |
| Immediate neighbours | guaD, ispA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 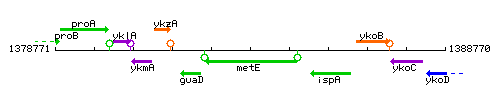
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed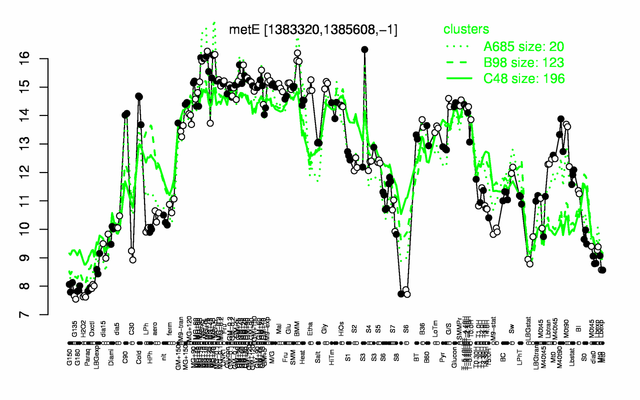
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU13180
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 5-methyltetrahydropteroyltri-L-glutamate + L-homocysteine = tetrahydropteroyltri-L-glutamate + L-methionine (according to Swiss-Prot)
- Protein family: vitamin-B12 independent methionine synthase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on ser/ thr/ tyr PubMed, S-cysteinylation after diamide stress (C719) PubMed
- Cys719 and Cys730 are S-bacillithiolated by NaOCl stress in B. subtilis and other Bacillus species PubMed PubMed
- MetE is generally most strongly S-bacillithiolated by NaOCl stress in B. subtilis and other Bacillus species PubMed PubMed
- Effectors of protein activity:
Database entries
- UniProt: P80877
- KEGG entry: [2]
- E.C. number: 2.1.1.14
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon: metE
- Regulation:
- Regulatory mechanism: S-box: transcription termination/ antitermination, the S-box riboswitch binds S-adenosylmethionine resulting in termination PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References