Difference between revisions of "CapB"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 66: | Line 62: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 84: | Line 78: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 118: | Line 112: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | * '''Regulatory mechanism:''' [[DegU]]-P: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/19734658 PubMed] | + | * '''Regulatory mechanism:''' |
| + | ** [[DegU]]-P: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/19734658 PubMed] | ||
| − | * '''Additional information:''' The operon is not expressed in the lab strain 168 due to lack of expression of [[DegQ]] resulting in inactivity of [[DegS]] and [[DegU]] {{PubMed|21965392}} | + | * '''Additional information:''' |
| + | ** The operon is not expressed in the lab strain 168 due to lack of expression of [[DegQ]] resulting in inactivity of [[DegS]] and [[DegU]] {{PubMed|21965392}} | ||
| + | ** expression of the operon is reduced in ''[[motA]]'' or ''[[motB]]'' mutants due to reduced [[DegU]] phosphorylation {{PubMed|24296669}} | ||
=Biological materials = | =Biological materials = | ||
| Line 144: | Line 141: | ||
<pubmed>20735481 </pubmed> | <pubmed>20735481 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | + | <pubmed>11751809,19420703, 19389763, 16233197 19734658 20564357 20564574 24296669 21965392</pubmed> | |
| − | <pubmed>11751809,19420703, 19389763, 16233197 19734658 20564357 20564574 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:12, 4 December 2013
- Description: capsular polyglutamate biosynthesis
| Gene name | capB |
| Synonyms | ywsC, pgsB |
| Essential | no |
| Product | poly-gamma-glutamate synthetase |
| Function | capsule synthesis |
| Gene expression levels in SubtiExpress: capB | |
| MW, pI | 43 kDa, 5.073 |
| Gene length, protein length | 1179 bp, 393 aa |
| Immediate neighbours | capC, rbsR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 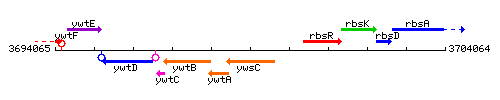
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
capsule biosynthesis and degradation
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35900
Phenotypes of a mutant
no synthesis of the poly-gamma-glutamate capsule PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: synthesis of poly-gamma-glutamate from L-Glu PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P96736
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Jia Mun Chan, Sarah B Guttenplan, Daniel B Kearns
Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis.
J Bacteriol: 2014, 196(4);740-53
[PubMed:24296669]
[WorldCat.org]
[DOI]
(I p)
Thi-Huyen Do, Yuki Suzuki, Naoki Abe, Jun Kaneko, Yoshifumi Itoh, Keitarou Kimura
Mutations suppressing the loss of DegQ function in Bacillus subtilis (natto) poly-γ-glutamate synthesis.
Appl Environ Microbiol: 2011, 77(23);8249-58
[PubMed:21965392]
[WorldCat.org]
[DOI]
(I p)
Tohru Kamei, Daisuke Yamashiro, Terumi Horiuchii, Yutaka Minouchi, Makoto Ashiuchi
Identification and biochemical characterization of membranous short-chain polyglutamate from Bacillus subtilis.
Chem Biodivers: 2010, 7(6);1563-72
[PubMed:20564574]
[WorldCat.org]
[DOI]
(I p)
Chuan-Mei Yeh, Jyh-Perng Wang, Shih-Ching Lo, Wen-Chia Chan, Ming-Yi Lin
Chromosomal integration of a synthetic expression control sequence achieves poly-gamma-glutamate production in a Bacillus subtilis strain.
Biotechnol Prog: 2010, 26(4);1001-7
[PubMed:20564357]
[WorldCat.org]
[DOI]
(I p)
Taku Ohsawa, Kensuke Tsukahara, Mitsuo Ogura
Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in gamma-poly-glutamic acid synthesis.
Biosci Biotechnol Biochem: 2009, 73(9);2096-102
[PubMed:19734658]
[WorldCat.org]
[DOI]
(I p)
Keitarou Kimura, Lam-Son Phan Tran, Thi-Huyen Do, Yoshifumi Itoh
Expression of the pgsB encoding the poly-gamma-DL-glutamate synthetase of Bacillus subtilis (natto).
Biosci Biotechnol Biochem: 2009, 73(5);1149-55
[PubMed:19420703]
[WorldCat.org]
[DOI]
(I p)
Cecilia Osera, Giuseppe Amati, Cinzia Calvio, Alessandro Galizzi
SwrAA activates poly-gamma-glutamate synthesis in addition to swarming in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 7);2282-2287
[PubMed:19389763]
[WorldCat.org]
[DOI]
(P p)
Yuji Urushibata, Shinji Tokuyama, Yasutaka Tahara
Difference in transcription levels of cap genes for gamma-polyglutamic acid production between Bacillus subtilis IFO 16449 and Marburg 168.
J Biosci Bioeng: 2002, 93(2);252-4
[PubMed:16233197]
[WorldCat.org]
[DOI]
(P p)
Yuji Urushibata, Shinji Tokuyama, Yasutaka Tahara
Characterization of the Bacillus subtilis ywsC gene, involved in gamma-polyglutamic acid production.
J Bacteriol: 2002, 184(2);337-43
[PubMed:11751809]
[WorldCat.org]
[DOI]
(P p)