Difference between revisions of "DegU"
| Line 60: | Line 60: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
* defect in [[biofilm formation]] {{PubMed|20815827}}, this can be suppressed by DegU-independent expression of [[YuaB]] {{PubMed|21742882}} | * defect in [[biofilm formation]] {{PubMed|20815827}}, this can be suppressed by DegU-independent expression of [[YuaB]] {{PubMed|21742882}} | ||
| + | * the mutation suppresses the mucoid phenotype of ''[[motA]]'' or ''[[motB]]'' mutants due to reduced expression of the ''[[pgsB]]-[[pgsC]]-[[pgsA]]'' operon {{PubMed|24296669}} | ||
| + | |||
=== Database entries === | === Database entries === | ||
| Line 82: | Line 84: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylated by [[DegS]] on Asp-56 [http://www.ncbi.nlm.nih.gov/sites/entrez/1901568 PubMed], this modulates DNA-binding activity | * '''Modification:''' phosphorylated by [[DegS]] on Asp-56 [http://www.ncbi.nlm.nih.gov/sites/entrez/1901568 PubMed], this modulates DNA-binding activity | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 120: | Line 122: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=degU_3644607_3645296_-1 degU] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=degU_3644607_3645296_-1 degU] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
** ''[[degS]]'': [[SigA]] {{PubMed|1688843}} | ** ''[[degS]]'': [[SigA]] {{PubMed|1688843}} | ||
** ''[[degU]]'': [[SigA]] {{PubMed|18502860}} | ** ''[[degU]]'': [[SigA]] {{PubMed|18502860}} | ||
| Line 161: | Line 163: | ||
<pubmed>12471443,,</pubmed> | <pubmed>12471443,,</pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>17850253,8878039, 14563871,1901568,1321152,1459944, 18194340 18978066 10908654,18502860,10094672,19389763, 20815827 21742882 18414485,19420703,2428811, 7746142, 18502860, 19389763, 19416356, 19734658 1688843 20070525 18197985 15598897 12950930 17590234 8955341 22496484 22745669 23123903 21965392 24123822 </pubmed> | + | <pubmed>17850253,8878039, 14563871,1901568,1321152,1459944, 18194340 18978066 10908654,18502860,10094672,19389763, 20815827 21742882 18414485,19420703,2428811, 7746142, 18502860, 19389763, 19416356, 19734658 1688843 20070525 18197985 15598897 12950930 17590234 8955341 22496484 22745669 23123903 21965392 24123822 24296669</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:09, 4 December 2013
- Description: two-component response regulator, regulation of degradative enzyme and genetic competence, involved in activation of capsule biosynthetic operon expression (together with SwrAA), non-phosphorylated DegU is required for swarming motility PubMed
| Gene name | degU |
| Synonyms | sacU, iep |
| Essential | no |
| Product | two-component response regulator |
| Function | regulation of degradative enzyme and genetic competence |
| Gene expression levels in SubtiExpress: degU | |
| Interactions involving this protein in SubtInteract: DegU | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 25 kDa, 5.602 |
| Gene length, protein length | 687 bp, 229 aa |
| Immediate neighbours | yviA, degS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 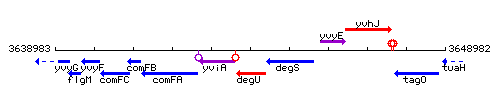
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biofilm formation, transcription factors and their control, phosphoproteins
This gene is a member of the following regulons
CcpA regulon, DegU regulon, TnrA regulon
The DegU regulon
The gene
Basic information
- Locus tag: BSU35490
Phenotypes of a mutant
- defect in biofilm formation PubMed, this can be suppressed by DegU-independent expression of YuaB PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants due to reduced expression of the pgsB-pgsC-pgsA operon PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activator of degradative enzyme genes (sacB and degQ) and of comK expression
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P13800
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The DegU regulon
Original Publications
Jia Mun Chan, Sarah B Guttenplan, Daniel B Kearns
Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis.
J Bacteriol: 2014, 196(4);740-53
[PubMed:24296669]
[WorldCat.org]
[DOI]
(I p)
Victoria L Marlow, Michael Porter, Laura Hobley, Taryn B Kiley, Jason R Swedlow, Fordyce A Davidson, Nicola R Stanley-Wall
Phosphorylated DegU manipulates cell fate differentiation in the Bacillus subtilis biofilm.
J Bacteriol: 2014, 196(1);16-27
[PubMed:24123822]
[WorldCat.org]
[DOI]
(I p)
Hiroshi Ishii, Teruo Tanaka, Mitsuo Ogura
The Bacillus subtilis response regulator gene degU is positively regulated by CcpA and by catabolite-repressed synthesis of ClpC.
J Bacteriol: 2013, 195(2);193-201
[PubMed:23123903]
[WorldCat.org]
[DOI]
(I p)
Fordyce A Davidson, Chung Seon-Yi, Nicola R Stanley-Wall
Selective heterogeneity in exoprotease production by Bacillus subtilis.
PLoS One: 2012, 7(6);e38574
[PubMed:22745669]
[WorldCat.org]
[DOI]
(I p)
Mitsuo Ogura, Kensuke Tsukahara
SwrA regulates assembly of Bacillus subtilis DegU via its interaction with N-terminal domain of DegU.
J Biochem: 2012, 151(6);643-55
[PubMed:22496484]
[WorldCat.org]
[DOI]
(I p)
Thi-Huyen Do, Yuki Suzuki, Naoki Abe, Jun Kaneko, Yoshifumi Itoh, Keitarou Kimura
Mutations suppressing the loss of DegQ function in Bacillus subtilis (natto) poly-γ-glutamate synthesis.
Appl Environ Microbiol: 2011, 77(23);8249-58
[PubMed:21965392]
[WorldCat.org]
[DOI]
(I p)
Adam Ostrowski, Angela Mehert, Alan Prescott, Taryn B Kiley, Nicola R Stanley-Wall
YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis.
J Bacteriol: 2011, 193(18);4821-31
[PubMed:21742882]
[WorldCat.org]
[DOI]
(I p)
Taryn B Kiley, Nicola R Stanley-Wall
Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation.
Mol Microbiol: 2010, 78(4);947-63
[PubMed:20815827]
[WorldCat.org]
[DOI]
(I p)
Mitsuo Ogura, Kensuke Tsukahara
Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP.
Mol Microbiol: 2010, 75(5);1244-59
[PubMed:20070525]
[WorldCat.org]
[DOI]
(I p)
Taku Ohsawa, Kensuke Tsukahara, Mitsuo Ogura
Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in gamma-poly-glutamic acid synthesis.
Biosci Biotechnol Biochem: 2009, 73(9);2096-102
[PubMed:19734658]
[WorldCat.org]
[DOI]
(I p)
Keitarou Kimura, Lam-Son Phan Tran, Thi-Huyen Do, Yoshifumi Itoh
Expression of the pgsB encoding the poly-gamma-DL-glutamate synthetase of Bacillus subtilis (natto).
Biosci Biotechnol Biochem: 2009, 73(5);1149-55
[PubMed:19420703]
[WorldCat.org]
[DOI]
(I p)
Monica Gupta, Kestur Krishnamurthy Rao
Epr plays a key role in DegU-mediated swarming motility of Bacillus subtilis.
FEMS Microbiol Lett: 2009, 295(2);187-94
[PubMed:19416356]
[WorldCat.org]
[DOI]
(I p)
Cecilia Osera, Giuseppe Amati, Cinzia Calvio, Alessandro Galizzi
SwrAA activates poly-gamma-glutamate synthesis in addition to swarming in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 7);2282-2287
[PubMed:19389763]
[WorldCat.org]
[DOI]
(P p)
Daniel T Verhamme, Ewan J Murray, Nicola R Stanley-Wall
DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis.
J Bacteriol: 2009, 191(1);100-8
[PubMed:18978066]
[WorldCat.org]
[DOI]
(I p)
Ayako Yasumura, Sadanobu Abe, Teruo Tanaka
Involvement of nitrogen regulation in Bacillus subtilis degU expression.
J Bacteriol: 2008, 190(15);5162-71
[PubMed:18502860]
[WorldCat.org]
[DOI]
(I p)
Jan-Willem Veening, Oleg A Igoshin, Robyn T Eijlander, Reindert Nijland, Leendert W Hamoen, Oscar P Kuipers
Transient heterogeneity in extracellular protease production by Bacillus subtilis.
Mol Syst Biol: 2008, 4;184
[PubMed:18414485]
[WorldCat.org]
[DOI]
(I p)
Kensuke Tsukahara, Mitsuo Ogura
Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB.
BMC Microbiol: 2008, 8;8
[PubMed:18197985]
[WorldCat.org]
[DOI]
(I e)
Kensuke Tsukahara, Mitsuo Ogura
Characterization of DegU-dependent expression of bpr in Bacillus subtilis.
FEMS Microbiol Lett: 2008, 280(1);8-13
[PubMed:18194340]
[WorldCat.org]
[DOI]
(P p)
Kazuo Kobayashi
Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2007, 66(2);395-409
[PubMed:17850253]
[WorldCat.org]
[DOI]
(P p)
Daniël T Verhamme, Taryn B Kiley, Nicola R Stanley-Wall
DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis.
Mol Microbiol: 2007, 65(2);554-68
[PubMed:17590234]
[WorldCat.org]
[DOI]
(P p)
Kana Shimane, Mitsuo Ogura
Mutational analysis of the helix-turn-helix region of Bacillus subtilis response regulator DegU, and identification of cis-acting sequences for DegU in the aprE and comK promoters.
J Biochem: 2004, 136(3);387-97
[PubMed:15598897]
[WorldCat.org]
[DOI]
(P p)
Leif Steil, Tamara Hoffmann, Ina Budde, Uwe Völker, Erhard Bremer
Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity.
J Bacteriol: 2003, 185(21);6358-70
[PubMed:14563871]
[WorldCat.org]
[DOI]
(P p)
Mitsuo Ogura, Kana Shimane, Kei Asai, Naotake Ogasawara, Teruo Tanaka
Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1685-97
[PubMed:12950930]
[WorldCat.org]
[DOI]
(P p)
L W Hamoen, A F Van Werkhoven, G Venema, D Dubnau
The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2000, 97(16);9246-51
[PubMed:10908654]
[WorldCat.org]
[DOI]
(P p)
C Fabret, V A Feher, J A Hoch
Two-component signal transduction in Bacillus subtilis: how one organism sees its world.
J Bacteriol: 1999, 181(7);1975-83
[PubMed:10094672]
[WorldCat.org]
[DOI]
(P p)
M Ogura, T Tanaka
Bacillus subtilis DegU acts as a positive regulator for comK expression.
FEBS Lett: 1996, 397(2-3);173-6
[PubMed:8955341]
[WorldCat.org]
[DOI]
(P p)
J Hahn, A Luttinger, D Dubnau
Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis.
Mol Microbiol: 1996, 21(4);763-75
[PubMed:8878039]
[WorldCat.org]
[DOI]
(P p)
B J Haijema, L W Hamoen, J Kooistra, G Venema, D van Sinderen
Expression of the ATP-dependent deoxyribonuclease of Bacillus subtilis is under competence-mediated control.
Mol Microbiol: 1995, 15(2);203-11
[PubMed:7746142]
[WorldCat.org]
[DOI]
(P p)
K Mukai, M Kawata-Mukai, T Tanaka
Stabilization of phosphorylated Bacillus subtilis DegU by DegR.
J Bacteriol: 1992, 174(24);7954-62
[PubMed:1459944]
[WorldCat.org]
[DOI]
(P p)
M K Dahl, T Msadek, F Kunst, G Rapoport
The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis.
J Biol Chem: 1992, 267(20);14509-14
[PubMed:1321152]
[WorldCat.org]
(P p)
M K Dahl, T Msadek, F Kunst, G Rapoport
Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase.
J Bacteriol: 1991, 173(8);2539-47
[PubMed:1901568]
[WorldCat.org]
[DOI]
(P p)
T Msadek, F Kunst, D Henner, A Klier, G Rapoport, R Dedonder
Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU.
J Bacteriol: 1990, 172(2);824-34
[PubMed:1688843]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes.
J Bacteriol: 1986, 168(1);380-8
[PubMed:2428811]
[WorldCat.org]
[DOI]
(P p)