Difference between revisions of "RecX"
(→References) |
|||
| Line 50: | Line 50: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
* reduced natural tranformation with plasmid or chromosomal DNA {{PubMed|23284295}} | * reduced natural tranformation with plasmid or chromosomal DNA {{PubMed|23284295}} | ||
| + | * drastically reduced survival of mature dormant spores after exposure to ultrahigh vacuum desiccation and ionizing radiation that induce single strand (ss) DNA nicks and double-strand breaks (DSBs) {{PubMed|24285298}} | ||
=== Database entries === | === Database entries === | ||
| Line 133: | Line 134: | ||
=References= | =References= | ||
| − | <pubmed>23284295 22383849 24285298 </pubmed> | + | <pubmed>23284295 22383849 24285298 24285298</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:21, 29 November 2013
- Description: recombination protein, modulates the SOS response and facilitates RecA-mediated recombinational repair and genetic recombination
| Gene name | recX |
| Synonyms | yfhG |
| Essential | no |
| Product | recombination protein |
| Function | DNA repair/ recombination |
| Gene expression levels in SubtiExpress: recX | |
| MW, pI | 30 kDa, 6.292 |
| Gene length, protein length | 792 bp, 264 aa |
| Immediate neighbours | yfhF, yfhH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 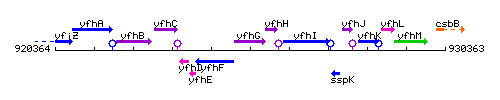
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed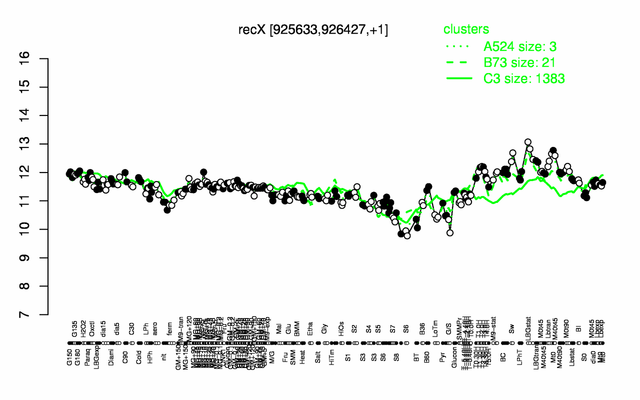
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU08520
Phenotypes of a mutant
- reduced natural tranformation with plasmid or chromosomal DNA PubMed
- drastically reduced survival of mature dormant spores after exposure to ultrahigh vacuum desiccation and ionizing radiation that induce single strand (ss) DNA nicks and double-strand breaks (DSBs) PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: recX family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31575
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References