Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' 4.5 S RNA<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''scr'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || small cytoplasmic RNA (scRNA, 4.5S RNA), |
| − | + | ||
| − | + | [[signal recognition particle]]-like ([[SRP]]) component | |
| − | |||
| − | |||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| ''' | + | |style="background:#ABCDEF;" align="center"|'''Function''' || presecretory protein translocation |
|- | |- | ||
| − | |style="background:# | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this RNA in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Scr Scr] |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yaaJ]]'', ''[[dnaX]]'' |
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|''' | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/scr_nucleotide.txt Gene sequence (+200bp) ]''' |
| + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/scr_protein.txt Protein sequence]''' | ||
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:scr_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| − | |||
| − | |||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <br/><br/><br/><br/><br/><br/> | |
| − | <br/><br/> | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[ | + | {{SubtiWiki category|[[protein secretion]]}}, |
| + | {{SubtiWiki category|[[essential genes]]}}, | ||
| + | {{SubtiWiki category|[[ncRNA]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 51: | Line 44: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * the gene is essential {{PubMed|7506707}} | ||
| + | === Database entries === | ||
| − | + | * '''DBTBS entry:''' no entry | |
| − | * ''' | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG00095] |
| − | * ''' | + | * '''Structure:''' |
=== Additional information=== | === Additional information=== | ||
| − | + | =The RNA= | |
| − | |||
| − | =The | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| Line 72: | Line 65: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | + | === Extended information on the RNA=== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | === Extended information on the | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | * ''' | + | * '''Kinetic information''' |
| − | * '''[[SubtInteract|Interactions]]:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** [[Scr]]-[[Ffh]] in the [[signal recognition particle]] | ||
| − | * '''[[Localization]] | + | * '''[[Localization]]''' |
=== Database entries === | === Database entries === | ||
| − | * '''Structure | + | * '''Structure''': |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Additional information=== | === Additional information=== | ||
| Line 108: | Line 82: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''''' [[yaaJ]]-[[scr]]'' | + | * '''Operon:''' ''[[yaaJ]]-[[scr]], [[scr]]'' |
| − | + | * '''Sigma factor:''' [[SigA]] (both promoters) | |
| − | |||
| − | * '''Sigma factor:''' [[SigA]] | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 118: | Line 90: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' transcribed as 354 nt precursor, cleaved at the 5' end by [[rnc| RNase III]] and at the 3' site by [[rnjA| RNase J1]] to yield a 275 nt molecule, final trimming to 271 nt sc RNA by [[rph| RNase PH]] [http://www.ncbi.nlm.nih.gov/sites/entrez/17576666 PubMed] |
=Biological materials = | =Biological materials = | ||
| Line 128: | Line 100: | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| − | + | =Labs working on this gene/protein= | |
| − | |||
| − | |||
| − | + | [[David Bechhofer]], Mount Sinai School, New York, USA [http://www.mountsinai.org/Research/Centers%20Laboratories%20and%20Programs/Bechhofer%20Laboratory?citype=Physician&ciid=Bechhofer%20David%20H%201255565 Homepage] | |
| − | |||
| − | = | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 140: | Line 108: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>9346318,2468993,17576666, 1372600,7506707 1383945 7538759 10224127 9677377 8662730 10222262</pubmed> |
| − | [[Category: | + | [[Category:small RNA]] |
Revision as of 14:54, 11 November 2013
- Description: 4.5 S RNA
| Gene name | scr |
| Synonyms | |
| Essential | yes |
| Product | small cytoplasmic RNA (scRNA, 4.5S RNA),
signal recognition particle-like (SRP) component |
| Function | presecretory protein translocation |
| Interactions involving this RNA in SubtInteract: Scr | |
| Immediate neighbours | yaaJ, dnaX |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 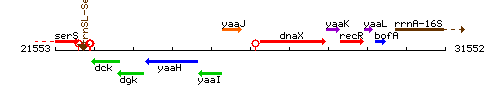
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
protein secretion, essential genes, ncRNA
This gene is a member of the following regulons
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
- the gene is essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
- Structure:
Additional information
The RNA
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
Extended information on the RNA
- Kinetic information
Database entries
- Structure:
Additional information
Expression and regulation
- Sigma factor: SigA (both promoters)
- Regulation:
- Regulatory mechanism:
- Additional information: transcribed as 354 nt precursor, cleaved at the 5' end by RNase III and at the 3' site by RNase J1 to yield a 275 nt molecule, final trimming to 271 nt sc RNA by RNase PH PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
Labs working on this gene/protein
David Bechhofer, Mount Sinai School, New York, USA Homepage
Your additional remarks
References