Difference between revisions of "LytC"
| Line 15: | Line 15: | ||
lysis, motility and general cell lysis induced by sodium azide | lysis, motility and general cell lysis induced by sodium azide | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU35620 lytC] |
lysis, motility and general cell lysis induced by sodium azide | lysis, motility and general cell lysis induced by sodium azide | ||
|- | |- | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[tuaA]]'', ''[[lytB]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[tuaA]]'', ''[[lytB]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU35620 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU35620 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU35620 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:lytC_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:lytC_context.gif]] | ||
Revision as of 14:00, 13 May 2013
- Description: N-acetylmuramoyl-L-alanine amidase, required for flagellar function PubMed
| Gene name | lytC |
| Synonyms | cwlB |
| Essential | no |
| Product | N-acetylmuramoyl-L-alanine amidase |
| Function | major autolysin, cell separation, wall turnover
lysis, motility and general cell lysis induced by sodium azide |
| Gene expression levels in SubtiExpress: lytC
lysis, motility and general cell lysis induced by sodium azide | |
| MW, pI | 52 kDa, 10.108 |
| Gene length, protein length | 1488 bp, 496 aa |
| Immediate neighbours | tuaA, lytB |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 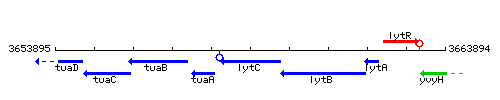
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed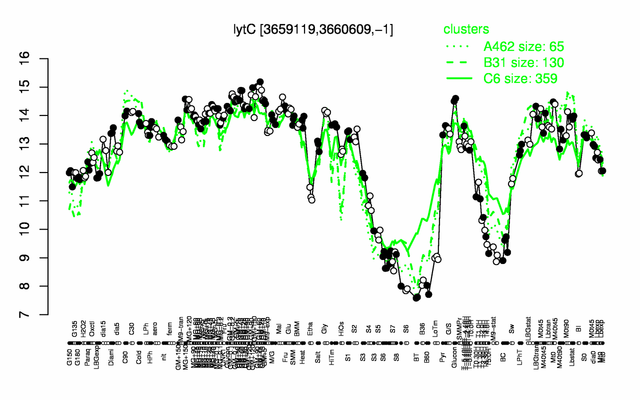
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, motility and chemotaxis
This gene is a member of the following regulons
SigD regulon, SlrR regulon, YvrHb regulon
The gene
Basic information
- Locus tag: BSU35620
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides (according to Swiss-Prot)
- Protein family: N-acetylmuramoyl-L-alanine amidase 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- secreted (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: Q02114
- KEGG entry: [3]
- E.C. number: 3.5.1.28
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yunrong Chai, Thomas Norman, Roberto Kolter, Richard Losick
An epigenetic switch governing daughter cell separation in Bacillus subtilis.
Genes Dev: 2010, 24(8);754-65
[PubMed:20351052]
[WorldCat.org]
[DOI]
(I p)
Rui Chen, Sarah B Guttenplan, Kris M Blair, Daniel B Kearns
Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity.
J Bacteriol: 2009, 191(18);5775-84
[PubMed:19542270]
[WorldCat.org]
[DOI]
(I p)
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
Hiroki Yamamoto, Shin-ichirou Kurosawa, Junichi Sekiguchi
Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases.
J Bacteriol: 2003, 185(22);6666-77
[PubMed:14594841]
[WorldCat.org]
[DOI]
(P p)
T Shida, H Hattori, F Ise, J Sekiguchi
Overexpression, purification, and characterization of Bacillus subtilis N-acetylmuramoyl-L-alanine amidase CwlC.
Biosci Biotechnol Biochem: 2000, 64(7);1522-5
[PubMed:10945275]
[WorldCat.org]
[DOI]
(P p)
P Margot, V Lazarevic, D Karamata
Effect of the SinR protein on the expression of the Bacillus subtilis 168 lytABC operon.
Microb Drug Resist: 1996, 2(1);119-21
[PubMed:9158733]
[WorldCat.org]
[DOI]
(P p)
V Lazarevic, P Margot, B Soldo, D Karamata
Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier.
J Gen Microbiol: 1992, 138(9);1949-61
[PubMed:1357079]
[WorldCat.org]
[DOI]
(P p)