Difference between revisions of "WapA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' cell wall-associated protein precursor <br/><br/> | + | * '''Description:''' cell wall-associated protein precursor, contact-dependent growth inhibition protein <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || cell wall-associated protein precursor | |style="background:#ABCDEF;" align="center"| '''Product''' || cell wall-associated protein precursor | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || intercellular competition |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU39230 wapA] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU39230 wapA] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/WapA WapA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 258 kDa, 6.131 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 258 kDa, 6.131 | ||
| Line 37: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[cell wall/ other]]}} | + | {{SubtiWiki category|[[cell wall/ other]]}}, |
| + | {{SubtiWiki category|[[toxins, antitoxins and immunity against toxins]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 82: | Line 81: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' cell wall binding domain as retention signal | + | * '''Domains:''' |
| + | ** cell wall binding domain as retention signal | ||
| + | ** C-terminal toxic domain with RNase activity (cleaves tRNAs) {{PubMed|23572593}} | ||
* '''Modification:''' | * '''Modification:''' | ||
| Line 89: | Line 90: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| + | ** [[YxxG]] inhibits the toxic activity of [[WapA]] {{PubMed|23572593}} | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** [[WapA]]-[[YxxG]] {{PubMed|23572593}} | ||
| − | * '''[[Localization]]:''' extracellular (signal peptide), cell wall binding domain as retention signal, major constituent of the secretome [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | + | * '''[[Localization]]:''' |
| + | ** extracellular (signal peptide), cell wall binding domain as retention signal, major constituent of the secretome [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | ||
=== Database entries === | === Database entries === | ||
| Line 143: | Line 147: | ||
=References= | =References= | ||
| − | <pubmed>11987133,16672620,18957862 16306698, 9537385 21821766</pubmed> | + | <pubmed>11987133,16672620,18957862 16306698, 9537385 21821766 23572593 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:49, 19 April 2013
- Description: cell wall-associated protein precursor, contact-dependent growth inhibition protein
| Gene name | wapA |
| Synonyms | |
| Essential | no |
| Product | cell wall-associated protein precursor |
| Function | intercellular competition |
| Gene expression levels in SubtiExpress: wapA | |
| Interactions involving this protein in SubtInteract: WapA | |
| MW, pI | 258 kDa, 6.131 |
| Gene length, protein length | 7002 bp, 2334 aa |
| Immediate neighbours | yxxG, yxxF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 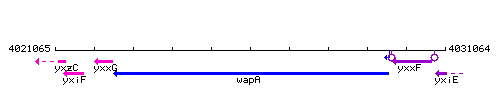
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall/ other, toxins, antitoxins and immunity against toxins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39230
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relexed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- cell wall binding domain as retention signal
- C-terminal toxic domain with RNase activity (cleaves tRNAs) PubMed
- Modification:
- Cofactor(s):
- Localization:
- extracellular (signal peptide), cell wall binding domain as retention signal, major constituent of the secretome PubMed
Database entries
- Structure:
- UniProt: Q07833
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References