Difference between revisions of "PcrA"
Raphael2215 (talk | contribs) |
|||
| Line 36: | Line 36: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 91: | Line 87: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | + | ** unwinding of the double-stranded ICEBs1 DNA is stimulated by [[HelP]] {{PubMed|23326247}} | |
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
** PcRA interacts with the [[RNA polymerase]] {{PubMed|21710567}} | ** PcRA interacts with the [[RNA polymerase]] {{PubMed|21710567}} | ||
| Line 118: | Line 114: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pcrA_719370_721589_1 pcrA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pcrA_719370_721589_1 pcrA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 151: | Line 147: | ||
==Original Publications== | ==Original Publications== | ||
'''Additional publications:''' {{PubMed|21710567}} | '''Additional publications:''' {{PubMed|21710567}} | ||
| − | <pubmed>12073041,9701819 ,16267290, 19943900 20723756 18276648 17449621 10899133 10199404 </pubmed> | + | <pubmed>12073041,9701819 ,16267290, 19943900 20723756 18276648 17449621 10899133 10199404 23326247 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:44, 13 February 2013
- Description: ATP-dependent DNA helicase, facilitates unwinding of ICEBs1 DNA for horizontal transfer
| Gene name | pcrA |
| Synonyms | yerF |
| Essential | yes PubMed |
| Product | ATP-dependent DNA helicase |
| Function | plasmid rolling-circle replication, conjugative transfer of ICEBs1 |
| Gene expression levels in SubtiExpress: pcrA | |
| Interactions involving this protein in SubtInteract: PcrA | |
| MW, pI | 83 kDa, 5.767 |
| Gene length, protein length | 2217 bp, 739 aa |
| Immediate neighbours | pcrB, ligA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 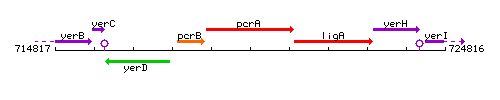
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed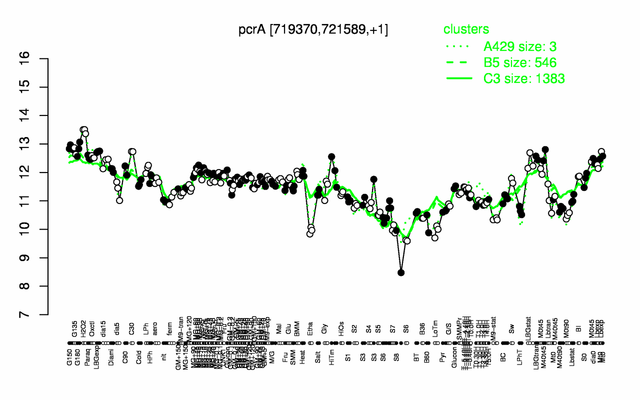
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, mobile genetic elements, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06610
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: uvrD-like helicase C-terminal domain (according to Swiss-Prot)
- Paralogous protein(s): YjcD
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
Database entries
- Structure: 2PJR (product complex, complex with sulphate ion, Geobacillus stearothermophilus), 1PJR (substrate complex, complex with DNA, Geobacillus stearothermophilus)
- UniProt: O34580
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Additional publications: PubMed