Difference between revisions of "Rnc"
(→Reviews) |
|||
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 59: | Line 55: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | * essential {{PubMed|23300471,12682299,11123676}} | |
| − | essential | + | * the ''[[rnc]]'' gene can be deleted in strains cured of the [[Skin element]] and [[prophage SPß]] (or upon deletion of the toxin genes ''[[txpA]]'' and ''[[yonT]]'') {{PubMed|23300471}} |
| − | |||
| − | ''rnc'' | ||
=== Database entries === | === Database entries === | ||
| Line 71: | Line 65: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 79: | Line 71: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' Endonucleolytic cleavage to 5'-phosphomonoester (according to Swiss-Prot) cleaves both 5'- and 3'-sites of the small cytoplasmic RNA precursor (''[[scr]]'') | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** Endonucleolytic cleavage of double-stranded RNA to 5'-phosphomonoester (according to Swiss-Prot) | ||
| + | ** cleaves both 5'- and 3'-sites of the small cytoplasmic RNA precursor (''[[scr]]'') | ||
| + | ** degradation of ''[[txpA]]'' mRNA-[[RatA]] RNA duplex (encoded on the [[Skin element]]) {{PubMed|23300471}} | ||
| + | ** degradation of ''[[yonT]]'' mRNA-[[as-YonT]] RNA duplex (encoded on the [[SP-beta prophage]]) {{PubMed|23300471}} | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| Line 119: | Line 115: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnc_1665710_1666459_1 rnc] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rnc_1665710_1666459_1 rnc] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|11021934}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|11021934}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 151: | Line 147: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed> 17194582 16179796 8936814 16855311 </pubmed> | + | <pubmed> 17194582 16179796 8936814 16855311 23300472 </pubmed> |
==Original publications== | ==Original publications== | ||
| − | <pubmed>9393702 11123676 7527387 9677377 17576666, 11021934 6409421 22123973 22412379 </pubmed> | + | <pubmed>9393702 11123676 7527387 9677377 17576666, 11021934 6409421 22123973 22412379 23300471 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:03, 10 January 2013
- Description: RNase III; required for the degradation of sense/antisense transcripts in Gram positive, low GC bacteria
| Gene name | rnc |
| Synonyms | rncS |
| Essential | yes |
| Product | endoribonuclease III |
| Function | cleaves both 5'- and 3'-sites of the small cytoplasmic RNA precursor (scr) |
| Gene expression levels in SubtiExpress: rnc | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 28 kDa, 8.076 |
| Gene length, protein length | 747 bp, 249 aa |
| Immediate neighbours | acpA, smc |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 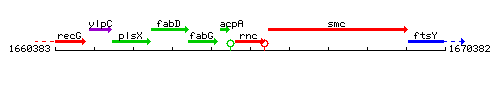
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
Rnases, translation, protein secretion, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15930
Phenotypes of a mutant
- essential PubMed
- the rnc gene can be deleted in strains cured of the Skin element and prophage SPß (or upon deletion of the toxin genes txpA and yonT) PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Endonucleolytic cleavage of double-stranded RNA to 5'-phosphomonoester (according to Swiss-Prot)
- cleaves both 5'- and 3'-sites of the small cytoplasmic RNA precursor (scr)
- degradation of txpA mRNA-RatA RNA duplex (encoded on the Skin element) PubMed
- degradation of yonT mRNA-as-YonT RNA duplex (encoded on the SP-beta prophage) PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P51833
- KEGG entry: [3]
- E.C. number: 3.1.26.3
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
David Bechhofer, Mount Sinai School, New York, USA Homepage
Ciaran Condon, IBPC, Paris, France Homepage
Your additional remarks
References
Reviews
Original publications
Sylvain Durand, Laetitia Gilet, Ciarán Condon
The essential function of B. subtilis RNase III is to silence foreign toxin genes.
PLoS Genet: 2012, 8(12);e1003181
[PubMed:23300471]
[WorldCat.org]
[DOI]
(I p)
Sylvain Durand, Laetitia Gilet, Philippe Bessières, Pierre Nicolas, Ciarán Condon
Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs.
PLoS Genet: 2012, 8(3);e1002520
[PubMed:22412379]
[WorldCat.org]
[DOI]
(I p)
Iñigo Lasa, Alejandro Toledo-Arana, Alexander Dobin, Maite Villanueva, Igor Ruiz de los Mozos, Marta Vergara-Irigaray, Víctor Segura, Delphine Fagegaltier, José R Penadés, Jaione Valle, Cristina Solano, Thomas R Gingeras
Genome-wide antisense transcription drives mRNA processing in bacteria.
Proc Natl Acad Sci U S A: 2011, 108(50);20172-7
[PubMed:22123973]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, Joshua B Blaustein, David H Bechhofer
Processing of Bacillus subtilis small cytoplasmic RNA: evidence for an additional endonuclease cleavage site.
Nucleic Acids Res: 2007, 35(13);4464-73
[PubMed:17576666]
[WorldCat.org]
[DOI]
(I p)
M A Herskovitz, D H Bechhofer
Endoribonuclease RNase III is essential in Bacillus subtilis.
Mol Microbiol: 2000, 38(5);1027-33
[PubMed:11123676]
[WorldCat.org]
[DOI]
(P p)
Hiroshi Kakeshita, Akihiro Oguro, Reiko Amikura, Kouji Nakamura, Kunio Yamane
Expression of the ftsY gene, encoding a homologue of the alpha subunit of mammalian signal recognition particle receptor, is controlled by different promoters in vegetative and sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2000, 146 ( Pt 10);2595-2603
[PubMed:11021934]
[WorldCat.org]
[DOI]
(P p)
A Oguro, H Kakeshita, K Nakamura, K Yamane, W Wang, D H Bechhofer
Bacillus subtilis RNase III cleaves both 5'- and 3'-sites of the small cytoplasmic RNA precursor.
J Biol Chem: 1998, 273(31);19542-7
[PubMed:9677377]
[WorldCat.org]
[DOI]
(P p)
W Wang, D H Bechhofer
Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci.
J Bacteriol: 1997, 179(23);7379-85
[PubMed:9393702]
[WorldCat.org]
[DOI]
(P p)
S Mitra, D H Bechhofer
Substrate specificity of an RNase III-like activity from Bacillus subtilis.
J Biol Chem: 1994, 269(50);31450-6
[PubMed:7527387]
[WorldCat.org]
(P p)
A T Panganiban, H R Whiteley
Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA.
Cell: 1983, 33(3);907-13
[PubMed:6409421]
[WorldCat.org]
[DOI]
(P p)