Difference between revisions of "YngE"
| Line 36: | Line 36: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 90: | Line 86: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| + | **[[YngH]]-[[YngE]] {{PubMed|19935659}} | ||
| − | * '''Localization:'' | + | * '''[[Localization]]:'' |
=== Database entries === | === Database entries === | ||
| Line 112: | Line 109: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yngE_1949682_1951217_-1 yngE] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yngE_1949682_1951217_-1 yngE] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigE]] {{PubMed|15699190,12662922}} | + | * '''[[Sigma factor]]:''' [[SigE]] {{PubMed|15699190,12662922}} |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 20:01, 15 December 2012
- Description: methylcrotonoyl-CoA carboxylase, subunit
| Gene name | yngE |
| Synonyms | |
| Essential | no |
| Product | methylcrotonoyl-CoA carboxylase, subunit |
| Function | mother cell metabolism, leucine utilization |
| Gene expression levels in SubtiExpress: yngE | |
| Interactions involving this protein in SubtInteract: YngE | |
| Gene length, protein length | 1785 bp, 595 aa |
| Immediate neighbours | nrnB, yngF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 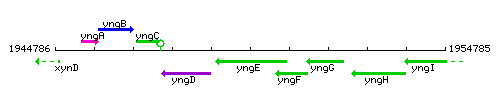
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed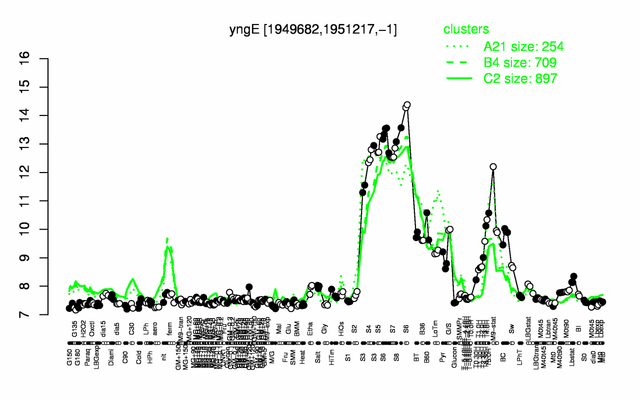
| |
Contents
Categories containing this gene/protein
utilization of amino acids, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU18210
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 3-methylbut-2-enoyl-CoA + Biotin-CO2 ---> 3-methyl-glutaconyl-CoA + biotin PubMed
- Protein family:
- Paralogous protein(s): YqjD
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31825
- KEGG entry: [3]
- E.C. number: 6.4.1.4
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tzu-Lin Hsiao, Olga Revelles, Lifeng Chen, Uwe Sauer, Dennis Vitkup
Automatic policing of biochemical annotations using genomic correlations.
Nat Chem Biol: 2010, 6(1);34-40
[PubMed:19935659]
[WorldCat.org]
[DOI]
(I p)
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Shane T Jensen, Erin M Conlon, Christiaan van Ooij, Jessica Silvaggi, José Eduardo González-Pastor, Masaya Fujita, Sigal Ben-Yehuda, Patrick Stragier, Jun S Liu, Richard Losick
The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis.
J Mol Biol: 2003, 327(5);945-72
[PubMed:12662922]
[WorldCat.org]
[DOI]
(P p)