Difference between revisions of "WprA"
| Line 14: | Line 14: | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU10770 wprA] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU10770 wprA] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/WprA WprA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 96 kDa, 9.58 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 96 kDa, 9.58 | ||
| Line 65: | Line 67: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** important for [[TatAY]]-[[TatCY]]-dependent export of [[YkuE]] and [[YwbN]] {{PubMed|23180473}} | ||
* '''Protein family:''' peptidase S8 family (according to Swiss-Prot) | * '''Protein family:''' peptidase S8 family (according to Swiss-Prot) | ||
| Line 83: | Line 86: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** [[WprA]]-[[TatAY]] {{PubMed|23180473}} | ||
| + | ** [[WprA]]-[[TatCY]] {{PubMed|23180473}} | ||
| − | * '''[[Localization]]:''' | + | * '''[[Localization]]:''' |
| + | ** extracellular (signal peptide) [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | ||
| + | ** membrane [http://www.ncbi.nlm.nih.gov/pubmed/18763711 PubMed] | ||
| + | ** cell wall {{PubMed|23180473}} | ||
=== Database entries === | === Database entries === | ||
| Line 104: | Line 112: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=wprA_1153789_1156473_1 wprA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=wprA_1153789_1156473_1 wprA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|22900538}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|22900538}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 137: | Line 145: | ||
=References= | =References= | ||
| − | <pubmed> 22900538 22923395 </pubmed> | + | <pubmed> 22900538 22923395 23180473</pubmed> |
<big>''Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J'' </big> | <big>''Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J'' </big> | ||
<big>'''RNA processing in ''Bacillus subtilis'': identification of targets of the essential RNase Y.''' </big> | <big>'''RNA processing in ''Bacillus subtilis'': identification of targets of the essential RNase Y.''' </big> | ||
Revision as of 09:13, 30 November 2012
- Description: secreted quality control protease
| Gene name | wprA |
| Synonyms | yisM |
| Essential | no |
| Product | secreted quality control protease |
| Function | protein quality control |
| Gene expression levels in SubtiExpress: wprA | |
| Interactions involving this protein in SubtInteract: WprA | |
| MW, pI | 96 kDa, 9.58 |
| Gene length, protein length | 2682 bp, 894 aa |
| Immediate neighbours | yisL, yisN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 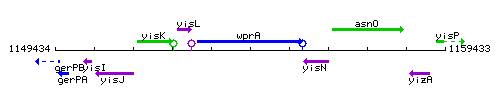
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed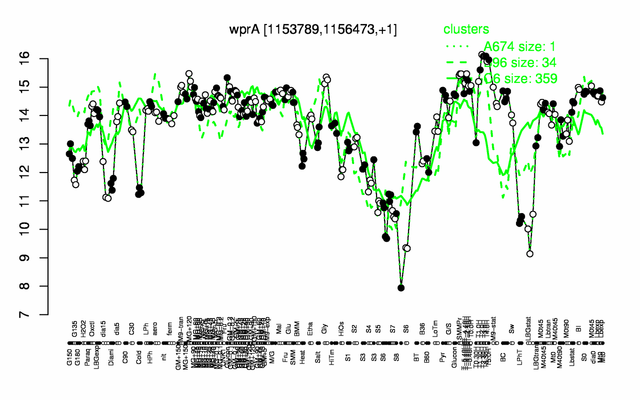
| |
Contents
Categories containing this gene/protein
cell wall/ other, proteolysis,
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU10770
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase S8 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54423
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- An antisense RNA is predicted forwprA PubMed
- the amount of the mRNA is substantially decreased upon depletion of RNase Y PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Carmine G Monteferrante, Calum MacKichan, Elodie Marchadier, Maria-Victoria Prejean, Rut Carballido-López, Jan Maarten van Dijl
Mapping the twin-arginine protein translocation network of Bacillus subtilis.
Proteomics: 2013, 13(5);800-11
[PubMed:23180473]
[WorldCat.org]
[DOI]
(I p)
Laxmi Krishnappa, Carmine G Monteferrante, Jan Maarten van Dijl
Degradation of the twin-arginine translocation substrate YwbN by extracytoplasmic proteases of Bacillus subtilis.
Appl Environ Microbiol: 2012, 78(21);7801-4
[PubMed:22923395]
[WorldCat.org]
[DOI]
(I p)
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
K Stephenson, C L Jensen, S T Jørgensen, C R Harwood
Simultaneous inactivation of the wprA and dltB genes of Bacillus subtilis reduces the yield of alpha-amylase.
Lett Appl Microbiol: 2002, 34(6);394-7
[PubMed:12028417]
[WorldCat.org]
[DOI]
(P p)
Haike Antelmann, Hiroki Yamamoto, Junichi Sekiguchi, Michael Hecker
Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach.
Proteomics: 2002, 2(5);591-602
[PubMed:11987133]
[WorldCat.org]
[DOI]
(P p)
J H Hageman, R L Switzer
ISP-4 and CWBP52 are proteins encoded by the same gene in Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 2);281
[PubMed:10075409]
[WorldCat.org]
[DOI]
(P p)
K Stephenson, C R Harwood
Influence of a cell-wall-associated protease on production of alpha-amylase by Bacillus subtilis.
Appl Environ Microbiol: 1998, 64(8);2875-81
[PubMed:9687444]
[WorldCat.org]
[DOI]
(P p)
P Margot, D Karamata
The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease.
Microbiology (Reading): 1996, 142 ( Pt 12);3437-44
[PubMed:9004506]
[WorldCat.org]
[DOI]
(P p)